DirectDicomImport¶
- MLModule¶
Purpose¶
DirectDicomImport is a MeVisLab module whose primary purpose is the import of DICOM and non-DICOM (image) objects. These are grouped, sorted, split, and composed to a MultiFileVolumeList under constraints defined as module parameters.
In DirectDicomImport the import of DICOM files is performed via a call to the Dicom Processor Library (DPL) and result MultiFileVolumes can be passed to other MeVisLab modules without duplicating the original data or needing an intermediate image representation.
Also many other image formats are imported, and - as well as DICOM files - are composed to volumes according to the DICOM tags or to the selected settings in the module parameters. See below for a list of supported formats.
Clicking in the Volumes list selects the output volume and double-clicking shows volume details.
Many options are available in Configuration window (opened with the Configuration button) to view, sort, analyze, and cache the generated image volumes and to pass any of them to the ML module output.
Output images are loaded on demand from the DICOM frames, and their content is not duplicated in an intermediate format. Thus pixel data is only read from DICOM frames when needed which reduce memory load.
Specialized FileListFilterPlugin modules can be connected to the Base input inputFilterPlugin to (pre-)filter and classify files, DICOM trees and tags before importing them.
DicomConfigurableMessageFilter modules can be connected to the Base input inputMessageFilterPlugin to suppress logging of import errors or warnings. Especially if the application or module network already handles issues, it can be safe and beneficial to filter messages in order to focus on important and non-handled ones.
Specialized MultiFileVolumeList modules derived from MultiFileVolumeListBaseOutput modules can be connected to the Base output to provide access to more than one image in the volume table at once.
Related documentation
DICOM in MeVisLabfor an overview about most DICOM functionality in MeVisLab.The alternative module
DicomImportfocusing more on DICOM only, fewer IODs, and speed.The command line tool MLABDumpTool for converting MeVisLab file formats which may be available with some deliveries of MeVisLab installers.
While this is the usage and parameter documentation of
DirectDicomImport, a more functional or architectural documentation can be found inFunctional Description of DirectDicomImport.For general import problems and error messages in
DirectDicomImportsee the Details section.Further references regarding DICOM in MeVisLab (may contain references to external or non-reachable documents):
Supported formats and backends
The currently supported formats and backends are listed below.
Important
Not all format versions can be read by all loaders.
Some file types can only be read if the files have the correct suffix and are part of the suffix list in
Explicit File Types.All non-built-in formats and backends could be excluded or disabled depending on QA-, license-, or installation contexts, and therefore they might not always be available.
The support of Transfer Syntax UIDs (see
Overview DICOM Support in MeVisLab - Transfer Syntax UIDs.2024.11.20.pdf), compressed data segments, and -files may vary depending on installed codecs and (seeMLDataCompressors).The support of compressed data segments and files may vary depending on installed codecs and DataCompressors (see
MLDataCompressorsandMLImageFormat).The support of DICOM character encodings may change between MeVisLab versions and depends on the current state of third party libraries which is beyond the scope of this document.
Since backends and libraries may change between MeVisLab releases the following list may vary and might not be not exhausting.
For DICOM files the following order of loaders is tested (if available):
Built-in DICOM importer of
DirectDicomImport
For non-DICOM files the following order of loaders is used (if available):
FileReaderPlugins installed in the ML Runtime Type System.
HistoLoad (usually not available in Public SDK or ADKs)
The following formats may be supported depending on installed backends and codecs:
.mlimage files loaded by
MLImageFormatLoadAll formats loaded by
ImageLoad(the list may be incomplete dependent on installed plugins):.tif, .tiff -Tagged Interface File Format
.dcm/.tif - DICOM/TIFF
LUMISYS
.pnm - PNM
Analyze
.png - PNG
.jpg - JPEG
All formats loaded by
itkImageFileReader(the list may be incomplete dependent on installed plugins):.img, .hdr, .img.gz - Analyze
.pic - BioRad
.bmp - Bitmap
.dcm, .dicom - Dicom
.gipl - Gipl
.jpg, .jpeg - JPEG
.lsm - Laser Scan Microscopy
.mha .mhd - MetaImage
.nrrd - Nearly Raw Raster Data
.nii - Nifti
.png - PNG
.spr - Stimulate
.tif, .tiff -Tagged Interface File Format
.vtk - VTK
Brains2MaskImageIO
GE4
GE5
IPL
MetaArray
SiemensVision
XML
Files loaded by installed FileReaderPlugins which currently are
DMFileReader
.dm3 - Digital Microfocal files, version 3
.dm4 - Digital Microfocal files, version 4
HistoFileReaderPlugin histological/pathological image formats (if HistoLoad module is available in the used installation)
.btf, .tif, .tiff - BigTiff including JPEG2000
.czi - ZEISS Microscopy
.dzi - DeepZoomImage
.jp2 - JPEG2000
.mi - MeVisLab proprietary multi-scale image format
.mrxs - MIRAX
.svs - Aperio
.wsv - Whole Slide Image
XYLibReaderPlugin (tried last)
.1, .2, .3, .4, .5, .6, .7, .8, .9 - Spectra Omicron/Leybold
.asc - text loader
.chi - ChiPLOT data
.cif - Powder Diffraction CIF
.cnf - Canberra CNF
.cpi - Sietronics Sieray CPI
.csv - comma-separated values
.dat - Rigaku DAT, RIET7/ILL_D1A5/PSI_DMC DAT, Canberra MCA, or text loader
.dbw - DBWS data file
.gss - gsas, Standard Powder data file
.mca - Canberra MCA
.neu - DBWS data file
.raw - Siemens/Bruker RAW
.rd - Philips PC-APD RD Raw Scan
.rit - DBWS data file
.sd - Philips PC-APD RD Raw Scan
.spe - Princeton Instruments WinSpec
.tab - comma-separated values
.tsv - comma-separated values
.txt - text loader
.udf - Philips UDF
.uxd - Siemens/Bruker Diffrac-AT UXD
.vms - VAMAS ISO-14976
.xdd - XFIT XDD
.xy - SPECS Specslab2 xy
Usage¶
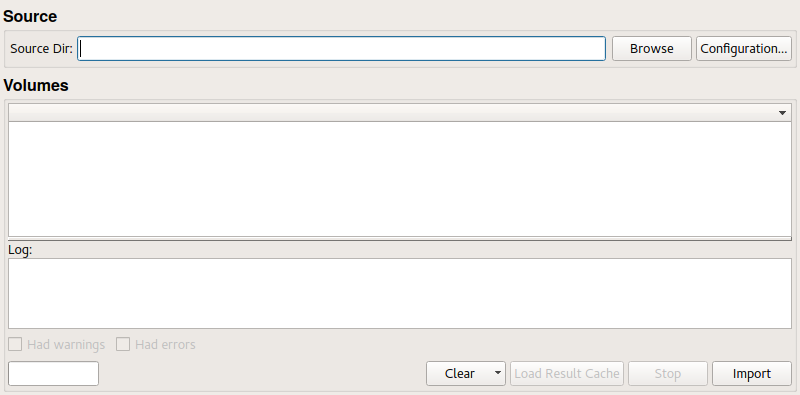
Select a source directory in the Source Dir or compose file/directory lists in Configuration -> Import Files or Configuration -> Import Directories.
Then use the Import button to parse the source files and directories. This composes image volumes from the DICOM files in it and shows them in the Volumes list.
Check the console and the Volumes list for issues and errors and other information and the generated image volumes.
Select any composed volume by clicking on it. The selected volume is made available at
output0if it is of an image type; possible overlay contents become available atoutput1.Or double-click a volume to open a window with volume details about the selected volume. Update
Volume Indexin details to update details to another volume if needed.Consider also connecting modules such as
MultiFileVolumeListIteratorOutputto process all or subsets of the volumes automatically, or to have multiple parallel outputs with modules such asMultiFileVolumeListImageOutput,MultiFileVolumeListSROutputor other modules derived fromMultiFileVolumeListBaseOutput.
Details¶
Performance issues¶
There are several reasons why importing can become slow, the most important and typical ones are listed here:
Problem: “A set of 100 MB DICOM files it imported in 1 second and another one of 300 MB takes 20 seconds. Why it is so much slower?”
DirectDicomImportmust load many parts of all DICOM files into memory to process and analyze their content. Since loading of this data is one of the most time consuming import steps a sophisticated caching strategy is used to optimize data reuse. If the cache, however, is too small to cache all imported files then multiply used data might not be cached and may have to be reloaded.Workarounds and solutions:
You may import the data in smaller steps, for example by importing one directory after another such that each imported chunk of data fits into the cache.
If you have enough main memory then consider increase
Max Tree Cached MBsto be some larger than the imported data and andMax Tag Cached MBsto have about a third part of the size ofMax Tree Cached MBs.
Problem: “One data of 100 MB DICOM files it imported in 1 second and another one of the same size is much slower. Where the difference may come from?”
Dependent of the content of the imported DICOM data
DirectDicomImportperform some analysis steps to finalize the result data. There are three steps which may become time consuming:Calculating the minimum and maximum voxel values in the image data. Most DICOM data files already provide these values as additional tag information (as smallest and largest pixel values tags) such that the
DirectDicomImportcan use them without further processing. However, some DICOM files do not provide them and then the default setting is to calculate them from the pixel data. This takes some time because all pixel data of those images have to be read and traversed.
Decomposing multiframe files. If
DirectDicomImportimports files of the newer enhanced multi-frame standard by decomposing them to single frame files to ensure correctly composed output volume. This costs time and temporary memory.Processing DICOM tags. The typical processing of DICOM tags in
DirectDicomImporthas an acceptable processing time, which, however, can degrade if there are large amounts of small tags or tags of huge sizes.Workarounds or solutions:
You may consider to disable the parsing of the image data (see
Min/Max Calculation), which, for example, might not be needed if you have aMinMaxScanin your processing pipeline anyway. However, if no min/max calculation is performed, then such a deactivating has the risk that output min/max values are much too large or even wrong which may affect or corrupt many operations in MeVisLab.You may disable the decomposition of multi-frame files, see
Decompose Multi Frame Filesfor details. Without this decomposition, however, there is the risk that the imported data has defects such as mirrored coordinate systems or incorrectly composed result volumes. Be aware of this before deactivating the decomposition.Some
DirectDicomImportversions provide a connector for DicomModifyTagPlugins such asDicomModifyTagsPluginor some more. These plugins can be used to modify or even remove these tags from the imported files before processing them. In the case of huge private tags this can speed up import performance significantly. The optionsCopy Private Top Level Source TagsandRemove private MFSQ tagcan be used to remove private tags from the generated outputs. However, take in consideration that the removed tag information will not stored in the generated DICOM tag trees any more and will be lost in further processing steps.
Trouble Shooting¶
Problem: “I get several messages in
Log, and I know what I do, or I have handled them all (e.g. with a connectedApplyDicomPixelModifiersmodule). Can I get rid of them?”Yes, you can get rid of nearly all messages: connect a
DicomConfigurableMessageFiltertoinputMessageFilterPluginand disable the handled ones, or those which are really are not of importance.However, be aware that the messages try to warn you about important errors or that data may not be in the state as you expect. By suppressing the messages with
DicomConfigurableMessageFilteryou will not get them any more. Hence use this option with care!An expected DICOM tag does not appear in
Tag Dumpor the tag(dump) does not match the original input data.The
Tag Dumpusually does not show the DICOM information of the originally imported and/or composed DICOM frames any more, but is a composed DICOM tree built from all input frames. It is a the MeVisLab-internal Structured Multi-Frame DICOM tree which represents the information of all input frames. If expected tags do not appear any more this usually has one of the following causes:Either the
Tag Dump Sizefield is not large enough. This is usually indicated at the end ofTag Dumpwith the message “tag list shortened and not shown completely” . Use a filter expression inFilterto filter explicitly for this tag or consider increasingTag Dump Size.A tag from the input frame has been removed, overwritten or added in the import process or due to configuration settings. The need for this behaviour can easily be understood regarding the value of the tag Number Of Frames. It must be revised if many single frame files are composed to a set of multiple frames to maintain the consistency of the composed DICOM tree.
Meta data information of non DICOM file(s) is displayed. Its format and content fully depends on the plugin which is used to load that data.
For displaying the original and unmodified DICOM information of a specific file use the
DicomTreeInfomodule.
Console message: “It was not possible to create valid image properties for this volume.”
This message often indicates that
DirectDicomImportcannot import the DICOM frame or volume, becauseit is of a type or modality which is still not or only partially supported,
the file is a manipulated or non standard DICOM file without pixel, spectroscopy, or deformation field data,
or the DICOM file/frame is invalid (often a subsequent problem of corrupted DICOM files as described below in the next issue).
The second case can happen if file pairs of the typical MeVisLab DCM/TIF file format are separated or incorrectly modified or renamed. The DICOM header file cannot be loaded correctly without the corresponding .tif file.
Stream output messages in the MeVisLab console or in the console of
DirectDicomImport.If you experience these messages then there is usually a problem in the imported DICOM data itself. Most of these messages comes from loader backends used to load the data, typically from the dcmtk library or even from a JPEG library used by dcmtk to decode DICOM pixel data. Often these messages indicate incorrect import processes, wrong volume compositions or even no or incompletely imported data.
Many of these message are caused by inconsistent DICOM tags, such as
unexpected or still unsupported values,
corrupted DICOM files or frames,
DICOM standard violations,
too short tag data sections,
unexpected or non standard value representations,
non ascending tag order
and so on.
Unfortunately
DirectDicomImportcannot correct or handle such errors, because in most of these cases the DICOM data itself is corrupted or it is not standard compliant.Reasons: Very typical reasons for this are
the transfer of DICOM files/frames via ftp in ASCII mode which replaces data bytes with new-line data bytes in binary DICOM data. Especially if only a few DICOM frames of a large set cause such errors, then this reason is very probable.
the transfer of DICOM files/frames via the www, via archive files or self-burned or old CDs or DVDs without check sum verification. Experiences with many DICOM file sources shows that due to the large data files these transfers tend to be unsafe.
Indicators: Check file sizes and dates in the folder with DICOM frames. In most cases (not always!) all DICOM frames have the same or nearly the same file size and only very small changes in its creation time. Indicators for corrupted files then might be
too small file sizes
irregular file sizes
irregular file dates
while the file names are in regular order.
Solutions: If you suspect any of the upper problems then try to get a new copy of the data after checking carefully where it comes from, how it was transferred, after adding checksums and verifying/correcting the data transfer modes (especially in ftp transfers and using zip or other archives).
Console message: “ The volume has a photometric interpretation PALETTECOLOR, PALETTE COLOR, YBR_FULL, or MONOCHROME1. The image voxels will be read as they are without translating them to grey scale or colors values. Consider using the module :module:`ApplyDicomPixelModifiers` after importing the volume to transform the image data appropriately”
Note that this is not an error but an important hint in order to use the imported image data correctly. It indicates that the imported DICOM frame(s) make use of a color model or palette index colors which are not translated by
DirectDicomImportor MultiFileVolumeList modules to the typical color model(s) used in MeVisLab, because this would require a modification of the DICOM (pixel) data. As mentioned, consider using the moduleApplyDicomPixelModifiersand/orDicomPaletteLUTfor solutions and further information.Console message: “ The volume contains DICOM tags Rescale Intercept != 0, Rescale Slope != 1 or Dose Grid Scaling != 1. The pixel data has not been transformed according to these values. Consider using the module ApplyDicomPixelModifiers after importing the volume to transform the image data appropriately.”
Note that this is not an error but an important hint in order to use the imported image data correctly, reasons are very similar to the upper message “The volume has a photometric interpretation…” and the solution is the same: see
ApplyDicomPixelModifiersandDicomRescalefor solutions and further information.Console message: “Could not determine min/max values from all frames. Min/Max values in image properties may be incorrect.”
In some cases the minimum and maximum value of the output image cannot determined. This can happen either, because the DICOM frame data cannot be read or decoded (then other problems exist) or the
Min/Max Calculationis set to “Only From Dicom Tags”. Try to use “Automatic” mode to solve the problem. Note that this may speed down import in some cases.Console messages: “Cerr: W: Invalid SOS parameters for sequential JPEG” and “DcmItem: Length of element XYZ is odd”
These messages and other similar ones indicate that the data inside of DICOM tag or pixel data can either be corrupted or compressed or coded in a sloppy or unsafe way. Without a detailed analysis of the binary frame data it is not possibly to decide which one is the case and whether it results to broken data or not. It is recommended to exclude such data from critical applications. See also the section Stream output messages in the MeVisLab console or in the console of
DirectDicomImportabove for details.Detected an uncompressed 2D DICOM frame with pixel data size matching rows*columns*sizeof(dataType)…
This message usually is printed when DirectDicomImport recognizes incorrect DICOM tag information. Tags in the imported frames are wrongly coded and violate the DICOM standard.
This commonly happens if original DICOM frames have been post-processed with other tools than the original creator or modality. Unfortunately these post-processing tools often tend to be buggy and create DICOM files which are not standard compliant any more. The mostly appearing scenario is, that some software does not support multi-frame or enhanced multi-frame files and therefore another tool is used to decomposed to them to single frame files. If you experience this, then request the original data before it was processed. Inform the source site about this possible misbehavior of the tool and import the data with DirectDicomImport again which usually can handle (enhanced) multi-frame files.
I get a message which is not discussed here, how can I get further information?
There is a vast amount of different messages which cannot be discussed all here, unfortunately this is out of the scope of this document.
Check the documentation and settings of
DicomConfigurableMessageFilter, some additional messages are discussed or even explained there.If the message contains additional information such as a specific module name then check the help documentation of that module for more information.
If the message contains information about a related third-party library (e.g. “dcm” comes from the dcmtk library) or a specific file format, then a web search may provide more information.
If all this does not help you may consider asking in the MeVisLab Forum (see https://forum.mevislab.de/index.php?action=login), best if you can provide example data such that the MeVisLab team can reproduce the message.
DICOM information appended to the output volumes and handling of SOPInstanceUID values¶
DirectDicomImport does not only compose files to volumes, but in case of DICOM files, it usually also creates new DICOM information as tree-like meta information appended to the volume, visible in Tag Dump.
In DICOM, the SOPInstanceUID is used to uniquely identify and/or reference a specific DICOM file. Therefore the composed DICOM information uses the following rules in composed DICOM information to replace/update/preserve SOPInstanceUID values and tags.
Imported single DICOM (image) files which are not composed with other files or decomposed into parts or frames, get the original and unchanged DICOM information as meta data, i.e. the original SOPInstanceUID tag and value are preserved.
If more than one DICOM frame from different sources/files are composed to a volume, a new DCMTree is created and appended as meta data.
On root level of the DCMTree a new and unique SOPInstanceUID is set.
Frames tags which are identical in all composed frames are usually stored only once. They are located on the same tree level where also a private sequence tag with entries for each frame containing the frame specific tags.
Frame specific information is stored in a private sequence tag under the private creator MeVis StructuredMF data with group id 0x0041 such as
... (0008,0018) UI 1 [SOPInstanceUID]:1.3.6.1.4.1.34261.18121618296094.1279.1561021778.1 ... (0041,0010) LO 1 [PrivateCreator]:MeVis StructuredMF data (0041,1001) US 1 [Private Tag: Element offset for number of dimensions, MeVis, MeVis StructuredMF data]:25 (0041,1002) SQ 0 [Private Tag: Element offset for dimension sequence, MeVis, MeVis StructuredMF data]:<Sequence tag. 25 sequenceItems, 0 values> Sequence Item 0: Frame specific information from first frame. ... Sequence Item N: Frame specific information from last frame.
If a frame comes from a enhanced DICOM file/frame and was decomposed with enabled
Decompose Multi Frame Files:the frame itself usually does not have its own SOPInstanceUID. Nevertheless the SOPInstanceUID of its source file is stored as SourceSOPInstanceUIDOfDecomposedFrame (lower eight bits of element id is 1) and
the source index of the frame is stored as SourceIndexOfDecomposedFrame (counting starts at 1, not at 0) (lower eight bits of element id is 1)
Both are preserved as private tags with the private creator Fraunhofer MEVIS and group id 0x041 in the frame specific entry such as
... Sequence Item I: ... (0041,0010) LO 1 [PrivateCreator]:Fraunhofer MEVIS (0041,1001) UL 1 [Private Tag: SourceIndexOfDecomposedFrame, Fraunhofer MEVIS, Fraunhofer MEVIS]:22 (0041,1002) UI 1 [Private Tag: SourceSOPInstanceUIDOfDecomposedFrame, Fraunhofer MEVIS, Fraunhofer MEVIS]:1.3.12.2.1107.5.99.3.30000012062513241298400000000 ...
If a frame comes from a non-enhanced DICOM file/frame, then it usually has an original SOPInstanceUID which is preserved normally as part of the frame specific entry such as
... Sequence Item I: ... (0008,0018) UI 1 [SOPInstanceUID]:1.2.840.113619.2.353.6945.3202330.15732.1410848781.479 ...
Tips¶
Not all DICOM files (especially those ones without image data, with private tags or newer DICOM standards) can be read reliably.
Volume compositions from slices and source files is a non trivial task and depends on import parameters, on DICOM tags, scan parameters and many other factors.
DirectDicomImportuses heuristics from the Dicom Processor Library (DPL) for that, and those import tools can fail. Try other parameter combinations if volume composition is not as expected.Even if caching is activated the import process can take a while. This can be interrupted only at a few places during operation, because the DPL and processings do not allow interruptions at all points. So please be patient when trying to interrupt.
When single or multiple DICOM frames are imported and composed to a volume, then currently the page extent for the result volume is set to the extent of the composed frames. This can lead to memory trouble if large frames are composed in MeVisLab when using the generated volumes. In some cases the usage of a
ImagePropertyConvertmodule afterDirectDicomImportor MultiFileVolumeList modules can help to compensate this problem.When activating Show Icon Column in Table this can degrade performance significantly, especially if a large number of volumes is created during the import process or if many large volumes are inside the list. Disable this flag for maximum import performance.
Windows¶
Default Panel¶
Input Fields¶
inputFilterPlugin¶
- name: inputFilterPlugin, type: FileListFilterPluginBase(MLBase)¶
At this connector modules such as
FileListFilterPlugincan be connected to filter or label the files to be imported. Also specific DPL configurations and settings can be assigned to influence the imported volumes and file sets. Note that these modules can be cascaded for multiple (pre) filter steps.
inputMessageFilterPlugin¶
- name: inputMessageFilterPlugin, type: DicomConfigurableMessageFilter(MLBase)¶
In some situations is may be of interest to suppress messages (errors, or warnings) generated by the
DirectDicomImportsince the user is aware of their origin and meaning, and because they shadow other messages which might have a higher importance. For this purpose a plugin can be connected to this input which filters out specific messages, for example aDicomConfigurableMessageFilter. If this input is left open then the messages are generated according toLog Parting And Sorting InformationandVerbose Mode.
Output Fields¶
The module has three outputs, one for a currently selected image, its overlay information if available, and a Base output for modules normally derived from MultiFileVolumeListBaseOutput (such as MultiFileVolumeListImageOutput or MultiFileVolumeListIteratorOutput) to provide access to the entire volume list and/or selected volumes.
output0¶
- name: output0, type: Image¶
The module output makes the the currently selected image volume available as ML image if it is a valid one.
The source image data is loaded on demand from the original DICOM source files.
outputAccessConnector¶
- name: outputAccessConnector, type: MultiFileVolumeListRefCounted(MLBase)¶
The Base connector can be used to connect additional MultiFileVolumeList modules derived from
MultiFileVolumeListBaseOutput. Those modules can provide access to multiple volumes in theDirectDicomImportat once.For details see for example the documentation of
MultiFileVolumeListBaseOutput,MultiFileVolumeListImageOutput,MultiFileVolumeListPROutput, and as well as many similar modules referenced in the see also section.For accessing this object via scripting, see the Scripting Reference:
MLMultiFileVolumeListWrapper.
output1¶
- name: output1, type: Image¶
If the currently selected volume is of an image type and if it contains DICOM overlay information then this possibly will be provided at this output. Typically it can be displayed with a
SoView2DOverlayor similar modules on top of the viewer showing the image from output connector 0. If multiple overlays are available, then they will be composed with an OR operation, overlay pixels will have value 1, background pixels 0. This image at this connector will be invalid if no valid DICOM image volume is selected or it does not contain valid overlay information. Only overlays according to the current DICOM standard are displayed; overlays according to the retired DICOM standard are not displayed. See http://medical.nema.org/Dicom/2011/11_03pu.pdf, section C.9 for details. The overlay image data is calculated on demand from the input DICOM information.
Parameter Fields¶
Field Index¶
Visible Fields¶
Tag Dump Size¶
Dump Private Tag Values¶
- name: dumpPrivateTagValues, type: Bool, default: FALSE¶
If this flag is enabled, the tag dump will also list and decode private DICOM tag values as far as possible with registered tag decoders. This might be a bit less stable then disabling this flag. Enabling this tag also can lead to significantly larger tag dumps. See
Tag Dumpfor details.
Num shown binary bytes¶
Annotate¶
- name: annotate, type: Bool, default: FALSE¶
If enabled then tags are annotated with further descriptive information such as possible types (1=Mandatory, 1C=Conditionally Mandatory, 2=Mandatory, may have zero or one item, 2C=Conditionally Mandatory, but may be empty, 3=Optional) and possible meanings of the tag. Note that no context nor any DICOM SOP class analysis is performed for that information and therefore all possible and perhaps redundant meanings are shown. See also
Tag Dumpfor details since larger values usually increase the size of the dump.
Filter¶
- name: regExLineFilter, type: String¶
If not empty then the regular expression is used to filter all lines of
Tag Dump; see that field for more details. Examples:Expression
Description
.*
Matches all string/lines.
.*(0008,0032).*
Matches all lines containing this tag ID.
.*logo.*
Matches anything containing ‘logo’ (case sensitive)
.*[lL][oO][gG][oO].*
Matches anything containing ‘logo’ (case insensitive)
.*.[jJ][pP][gG]
Matches anything ending with .jpg where jpg is case insensitive.
.*.JPG|.*.jpg
Matches all paths ending with either ‘.jpg’ or with ‘.JPG’.
.*\d.*
Matches all paths containing at least one digit.
.*[[:digit:]].*
Same as above.
.*[[:xdigit:]].*
Matches all paths containing at least one hexadecimal digit.
.*\l.*
Matches all paths containing at least one lower case character.
.*[[:lower:]].*
Same as above.
.*\u.*
Matches all paths containing at least one upper case character.
.*[[:upper:]].*
Same as above.
.*\s.*
Matches all paths containing at least one white space character.
.*[[:space:]].*
Same as above.
.*[[:blank:]].*
Matches all paths containing at least one non-line-separating whitespace.
.*[[:word:]].*
Matches all paths containing at least one word character (alphanumeric characters plus the underscore).
.*[[:w:]].*
Same as above.
.*[[:graph:]].*
Matches all paths containing at least one graphical character.
.*[5-7].*
Matches all paths containing at least one of the digits 5, 6 or 7.
Tag Dump¶
- name: tagDump, type: String, persistent: no¶
Shows the DICOM tag list or the meta data of the currently selected volume. IMPORTANT: The meta data dump will be clamped if it is larger than
Tag Dump Sizeto avoid that creating and displaying it takes too long. Check An expected DICOM tag does not appear in Tag Dump or the tag(dump) does not match the original input data in trouble shooting section for solutions. Consider also to make use ofDicomTreeInfomodule to display the original and unmodified DICOM information of a specific file.
Full Source Dir Path¶
- name: fullPath, type: String, persistent: no¶
This is the absolute path of
Source Dirdirectory from where DICOM files are going to be imported. This field is often useful to connect that path to other fields for further usage or to feed in an absolute path when the GUI ofDirectDicomImportis not available (for example in code generated networks).
Input directory list¶
- name: fullUserSrcPaths, type: String¶
Line separated additional directory paths which are also imported after parsing
Full Source Dir Path. Important: Each specified directory is imported in a single import step which means that files from two directories specified in this field will never be composed to one volume. This can especially be useful to avoid that files from different directories are composed. See alsoInput file list.Newer versions of
DirectDicomImportresolve MeVisLab script variables such as $(TMPDIR) or $(DemoDataPath) if a script engine is available. Note that such variables will not be expanded ifDirectDicomImportruns inside a script-less environment (e.g. standalone c++-tests).
Use Full User Src Dir Paths Comment Prefix¶
- name: useFullUserSrcDirPathsCommentPrefix, type: Bool, default: FALSE¶
If activated, it enables non empty
Line comment prefixas comment at the begin of lines inInput directory listto make such lines ignored in imports.
Line comment prefix (fullUserSrcDirPathsCommentPrefix)¶
- name: fullUserSrcDirPathsCommentPrefix, type: String, default: ##¶
This is the string which can be used at begin of lines in
Input directory listto ignore them in imports; if this string is empty orUse Full User Src Dir Paths Comment Prefixis off, it will be ignored.
Full Cache Path¶
- name: fullCachePath, type: String, persistent: no¶
This read-only field (expanded from
Cache Path) shows the full path name used for storing and loading MultiFileVolumeList caches.
Use relative paths in cache files¶
- name: useRelativeCachePaths, type: Bool, default: FALSE, deprecated name: useRelativeCachePathsFld¶
If possible use relative paths in cache files. This, however, is only possible for files which are located next to the written cache file or in directories which are next to the cache file. All other paths will still be generated and stored as absolute paths.
Write cache files for each volume¶
- name: writeSingleVolumeCacheFiles, type: Bool, default: FALSE¶
If disabled then only one large cache file for all volumes is generated. If enabled then for each volume a single cache file is written as well as a parent cache file. This allows to load single volumes from their corresponding cache files without the need to all volumes at once.
Automatically create cache directory¶
- name: autoCreateCachePath, type: Bool, default: FALSE¶
Automatically create the cache directory if it does not yet exist.
Cache File Name¶
- name: cacheFileName, type: String, default: DDICacheFile¶
Name of cache file in which
DirectDicomImportcan store cache information to speed up file reloads etc. The ending ‘.mlDDICache’ will automatically be added. Actually the cache files are stored MultiFileVolumeLists and therefore can also be managed, loaded and displayed with modules derived fromMultiFileVolumeListBaseOutput.Note that this path is insensitive if
Use result cachingis disabled.
Log¶
- name: console, type: String, persistent: no¶
Shows logging, verbose and error information of the file parse and volume composition process.
Check this console for warnings and error after importing DICOM slices to see whether everything worked fine.
Console Size In Kilo Bytes¶
- name: consoleSizeInKiloBytes, type: Integer, default: 64¶
Logging outputs larger than this value will be clamped (to avoid performance loss due to too long logging outputs).
Clear Console¶
- name: clearConsole, type: Trigger¶
Clears the console output. This operation is available via the enumerator field Clear.
Volume Index¶
- name: outVolume, type: Integer, default: 0, minimum: 0, maximum: :field:`numVolumes`¶
This indexes the volume currently selected in the volume list and passed to the module output.
Dpl Import¶
Clear Dcm Tree Cache¶
- name: clearDcmTreeCache, type: Trigger¶
Clear cached DICOM tag trees. Usually only needed if contents of already cached files have changed. This operation is available via the enumerator field Clear.
Clear Result Cache¶
- name: clearResultCache, type: Trigger¶
Clear cached result volumes. This operation is available via the enumerator field Clear.
Clear Volume List¶
- name: clearVolumeList, type: Trigger¶
Makes the current volume list empty, that means imported volumes or loaded volumes from cached files are thrown away. Stored files on disk or stored cache files are not affected. This operation is available via the enumerator field Clear.
Load Result Cache¶
- name: loadResultCache, type: Trigger¶
Reloads cached result volumes.
Prefer cache reload before import¶
- name: preferCacheBeforeImport, type: Bool, default: FALSE¶
If enabled then before an import it is always tried to load a cache; only if that fails a normal import is performed. This flag is only used if
Use result cachingis enabled. Requires enabled result caching.
Save Result Cache¶
- name: saveResultCache, type: Trigger¶
Stores the current volumes list in a result cache. Requires enabled result caching.
Stop¶
- name: stopImporting, type: Trigger¶
If pressed the DICOM Processor Library (DPL) importing process is stopped as soon as possible (even that may take a while, please be patient!).
Clear After Import¶
- name: clearAfterImport, type: Bool, default: FALSE¶
If enabled then the DICOM tree cache is cleared after importing has finished.
Clear Single Frame Cache After Import¶
- name: clearSingleFrameCacheAfterImport, type: Bool, default: TRUE¶
If enabled (the recommended default) then the cache for decomposed multi frame trees is cleared automatically after importing has finished since they are normally not needed any more.
Modules such as
DicomModifyMultiFileVolumeExportrequire the decomposed frame information if they shall be exported to files after a finished imported process. In such casesClear Single Frame Cache After Importcan be disabled to make these frame decompositions available for further processing. Then the cache must be cleared withClear Single Frame Cachemanually to avoid wasting memory.
Clear Single Frame Cache¶
- name: clearSingleFrameCache, type: Trigger¶
If enabled then the cache for decomposed multi frames is cleared. Only enabled if
Clear Single Frame Cache After Importis disabled. This should typically be used if the decomposed frames are not needed any more, for example after exporting them with aDicomModifyMultiFileVolumeExport.
Progress Info¶
- name: progressInfo, type: String, persistent: no¶
Shows descriptive information about the state of the import progress.
Max Tree Cached MBs¶
- name: maxTreeCachedMBs, type: Integer, default: 1000, minimum: -1, deprecated name: maxCachedMBs¶
Mega bytes of memory spent for caching data of loaded DICOM trees, values < 0 means unlimited. Too small values can degrade performance, and high values can improve performance. Spend as much memory without endangering the stability of the application.
Max Tag Cached MBs¶
- name: maxTagCachedMBs, type: Integer, default: 500¶
Mega bytes of memory spent for caching data of loaded DICOM trees, values < 0 means unlimited. See also
Max Tree Cached MBs.
Edit DICOM cache size¶
- name: editDicomCacheSize, type: Bool, default: FALSE¶
Normally the internal data cache for loaded DICOM frames and tags should not be controlled by a specific
DirectDicomImportmodule, because it is a shared over the entire MeVisLab application. However, if an application needs control over the cache size used byDirectDicomImportmodules and therefore wants to modify the cache sizes then this flag can be enabled. Note that it is strongly recommended that only onDirectDicomImportinstance in an application or network should control the cache size to avoid side effects. SeeMax Tag Cached MBsandMax Tree Cached MBsfor further information.
Use result caching¶
- name: useResultCaching, type: Bool, default: FALSE¶
Traversed files and directories are cached for fast reload with
Load Result Cacheif this check box is enabled.
Num Volumes¶
- name: numVolumes, type: Integer, persistent: no¶
This read-only field shows how many volumes were composed and are shown in the volume table.
Total Slices¶
- name: totalSlices, type: Integer, persistent: no¶
This read-only field shows the sum of slices of all volumes shown in the volume table.
Total Time Points¶
- name: totalTimePoints, type: Integer, persistent: no¶
This read-only field shows the sum of time points of all volumes shown in the volume table.
Total Slices Mult Time Points¶
- name: totalSlicesMultTimePoints, type: Integer, persistent: no¶
This read-only field shows the sum of slices multiplied with the number of time points of all volumes shown in the volume table.
Total Voxels¶
- name: totalVoxels, type: Double, persistent: no¶
This read-only field shows the sum of number of voxels of all volumes shown in the volume table.
X¶
- name: extX, type: Integer, persistent: no¶
This read-only field shows the x-extent of the currently selected volume.
Y¶
- name: extY, type: Integer, persistent: no¶
This read-only field shows the y-extent of the currently selected volume.
Z¶
- name: extZ, type: Integer, persistent: no¶
This read-only field shows the z-extent of the currently selected volume.
C¶
- name: extC, type: Integer, persistent: no¶
This read-only field shows the color dimension extent of the currently selected volume.
T¶
- name: extT, type: Integer, persistent: no¶
This read-only field shows the time dimension extent of the currently selected volume.
U¶
- name: extU, type: Integer, persistent: no¶
This read-only field shows the u-dimension extent of the currently selected volume.
Num Vol Voxels¶
- name: numVolVoxels, type: Double, persistent: no¶
This read-only field shows number of voxels of the currently selected volume.
Volume Info Dump¶
- name: volumeInfoDump, type: String, persistent: no¶
Shows a collection of available information about the currently selected volume.
File List Info¶
- name: fileListInfo, type: String, persistent: no¶
Shows the list of files corresponding to the currently selected volume.
Suppressed Files and Suffixes¶
- name: suppressedFilesAndSuffixes, type: String, default: .mlab .db .log .lst .xml .rar .zip .img .exe .ini .inf .jxr .ndpi .svn-base .vmu entries all-wcprops .DDI-SMF-TreeCacheFile.dcm, deprecated name: suppressedSuffixes¶
This is a space separated list of suffixes or file names which are not taken into account while importing and composing files. All strings in this list not starting with “.” are taken as file name and must match exactly to suppress a file found in the import process. All strings starting with a “.” are considered as suffixes. The matching process is case insensitive, i.e. file.test will suppress file.test as well as FILE.TEST.
Explicit File Types¶
- name: explicitFileTypes, type: String, default: .bmp .btf .dm3 .dm4 .cnf .cpi .czi .dat .dzi .gipl .gsa .hdr .jp2 .jpg .jpeg .lsm .mhd .mi .mlimage .nhdr .nii .mca .mrxs .nrrd .pgm .pic .png .pnm .ppm .raw .rd .rek .spe .spr .svs .tif .tiff .txt .uxd .vms .vtk .wsv .xy¶
Files types imported as normal files without DICOM analysis.
Input file list¶
- name: inputFileList, type: String, deprecated name: i¶
A line separated list of absolute paths to files to be imported; see also
Input directory list. Newer versions ofDirectDicomImportresolve MeVisLab script variables such as $(TMPDIR) or $(DemoDataPath) if a script engine is available. Note that such variables will not be expanded ifDirectDicomImportruns inside a script-less environment (e.g. standalone c++-tests).
Use Input File List Comment Prefix¶
- name: useInputFileListCommentPrefix, type: Bool, default: FALSE¶
If activated, it enables non empty
Line comment prefixas comment at the begin of lines inInput file listto make such lines ignored in imports.
Line comment prefix (inputFileListCommentPrefix)¶
- name: inputFileListCommentPrefix, type: String, default: ##¶
This is the string which can be used at begin of lines in
Input file listto ignore them in imports; if this string is empty orUse Input File List Comment Prefixis off, it will be ignored.
Had Warnings¶
- name: hadWarnings, type: Bool, persistent: no¶
This flag will be checked if any warnings occurred, otherwise it will be cleared.
Had Errors¶
- name: hadErrors, type: Bool, persistent: no¶
This flag will be checked if any errors occurred, otherwise it will be cleared.
Log Dicom Cache Infos¶
- name: logDicomCacheInfos, type: Bool, default: FALSE¶
If enabled then DICOM cache usage is logged into the DDI console during import operations.
Warn Dicom Cache Overflows¶
- name: warnDicomCacheOverflows, type: Bool, default: FALSE¶
If enabled then overflows of the DICOM cache are logged into the MeVisLab console as warnings.
Volume Infos¶
- name: volumeInfos, type: String, persistent: no¶
This read-only field shows a table with much information about generated image volumes and allows their selection with mouse clicks or by using the Up or Down keys to change the current selection. The volume at the selected index (shown in the ‘Idx’ column’ of the Volumes list) is passed to the image output of
DirectDicomImport.Its columns are configured in
Column Configin the tab Configuration -> Volume List Options -> Columns.
Sort Mode¶
- name: sortMode, type: Enum, default: NumVoxels¶
This field selects the mode how the volume list is sorted in the Idx column. To see the right order in the volume table make the Idx column sorted in the table.
Values:
Title |
Name |
Description |
|---|---|---|
No Sort |
NoSort |
The volume list is displayed as it was generated during import. |
Valid Volumes |
ValidVolumes |
Sorting criteria is whether the volumes have valid image data. |
Num Bytes |
NumBytes |
The volume are sorted according to their number of bytes in image data. |
Num Voxels |
NumVoxels |
The volume are sorted according to their number of voxels in image data. |
XExtent |
XExtent |
The volume are sorted according to the spatial x-extent of their image data. |
YExtent |
YExtent |
The volume are sorted according to the spatial y-extent of their image data. |
ZExtent |
ZExtent |
The volume are sorted according to the spatial z-extent of their image data. |
CXExtent |
CXExtent |
The volume are sorted according to the color dimension extent of their image data. |
TExtent |
TExtent |
The volume are sorted according to the time extent of their image data. |
UExtent |
UExtent |
The volume are sorted according to the extent of user dimension of their image data. |
Voxel Volume |
VoxelVolume |
The volume are sorted according to the geometrical volume of the entire image data. |
Voxel Type Size |
VoxelTypeSize |
The volume are sorted according to the byte-size of the data type of their image data. |
Voxel Voxel Components |
VoxelVoxelComponents |
The volume are sorted according to the number of components of the data type of their image data. |
Num Slice Bytes |
NumSliceBytes |
The volume are sorted according to the number of bytes of a 2D x/y dimensional slice/frame in their image data. |
Num Slice Voxels |
NumSliceVoxels |
The volume are sorted according to the number of voxels of a 2D x/y dimensional slice/frame in their image data. |
Sort According Dcm Tag |
SortAccordingDcmTag |
Sorts according to the string value retrieved from a given DICOM tag. See |
Filter Label |
FilterLabel |
The volumes are sorted according to their volume label provided via a |
Sort Upwards¶
- name: sortUpwards, type: Bool, default: TRUE¶
Large values of the value specified in ‘Sort Mode’ are sorted to the start of the volume list if this flag is true, otherwise they are sorted to the end. Note that for correct sort order the Idx column in the table must be selected for sorting.
Idx Sort Tag Value¶
- name: idxSortTagValue, type: String, default: Modality¶
The string value of the DICOM tags with this tag id are used for sorting the volume list if
Sort Modeis Sort According Dcm Tag, otherwise this value is ignored and the field is insensitive or invisible. If tag value cannot be retrieved from the volume for any reason then an empty string is used for sorting. The tag id string must have a format like (ABCD, EFGH) where ABCD and EFGH are hexadecimal numbers describing 16 bit unsigned integers where ABCD are the high and EFGH are the low bits of the word. Sorting according tags whose values are numerical but represented as string is influenced by theUse Numeric Tag Value Sorting.
Use Numeric Tag Value Sorting¶
- name: useNumericTagValueSorting, type: Bool, default: FALSE¶
If enabled then tags with numeric string values are interpreted as sign 32 bit integers (and printed with “-” for negative values if necessary) and leading spaces to fill 8 characters to make all tag values sortable uniquely. If disabled then from the numeric string value tags the normal string values are read as they are stored in the tag. This flag is also used by sorting, see
Sort Modeif the mode Sort According Dcm Tag is used.
Thumbnail Extent¶
- name: thumbNailsExt, type: Integer, default: 40¶
Defines the extent (in pixels) of thumb nails shown in the volume list. Note that this field is insensitive if
Show icon column in tableis disabled.
Id (userColumnTag0)¶
- name: userColumnTag0, type: String¶
First tag Id of the tag whose value can be shown in a column in the volume list. The tag id string must have a format like (ABCD, EFGH) where ABCD and EFGH are hexadecimal numbers describing 16 bit unsigned integers where ABCD are the high and EFGH are the low bits of the word. See also
User Tag0where this column can be enabled.
Id (userColumnTag1)¶
- name: userColumnTag1, type: String¶
Second tag Id of the tag whose value can be shown in a column in the volume list. The tag id string must have a format like (ABCD, EFGH) where ABCD and EFGH are hexadecimal numbers describing 16 bit unsigned integers where ABCD are the high and EFGH are the low bits of the word. See also
User Tag1where this column can be enabled.
Id (userColumnTag2)¶
- name: userColumnTag2, type: String¶
Third tag Id of the tag whose value can be shown in a column in the volume list. The tag id string must have a format like (ABCD, EFGH) where ABCD and EFGH are hexadecimal numbers describing 16 bit unsigned integers where ABCD are the high and EFGH are the low bits of the word. See also
User Tag2where this column can be enabled.
Icon Build Mode¶
- name: iconBuildMode, type: Enum, default: UseMiddleSlice¶
Selects how/from which input slices the thumbnail icon in the volume table is generated from input image. Note that this field is insensitive if the checkbox Show Icon Column in Table is disabled.
Use First Slice Takes the first slice from the image to build the thumbnail.
Use Middle Slice Takes the middle slice from the image to build the thumbnail.
Important:
When activating Show Icon Column in Table this can degrate performance significantly, especially if a large number of volumes is created during the import process or if many large volumes are inside the list. Disable this flag for maximum import performance.
Show icon column in table¶
- name: icon, type: Bool, default: FALSE¶
If enabled then an overview icon of the volume is shown in a column in the volume list. The extent and the way how the icon is generated is defined in the fields
Thumbnail ExtentandIcon Build Mode. Note that this field is insensitive if the checkbox Show Icon Column in Table is disabled.
User Tag0¶
User Tag1¶
User Tag2¶
Column Config¶
- name: columnConfig, type: String, default: # Note: Comments must always be prefixed at line start with a '#'., , # Default setting:, validImage hasIssues fileType voxelType imgExt numVoxels numSlices numTimePoints numSrcFiles modality patientID patientSex patientsName studyID, , #modality studyDate acquisitionTime numSlices spacingBetweenSlices studyDescription seriesDescription acquisitionDate imageComments patientsName patientSex patientsBirthDate patientID referringPhysiciansName admittingDiagnosesDescription, , # Available values are:, #userTag0 userTag1 userTag2 validImage hasIssues importTime fileType voxelType imgExt voxelExtent numVoxels numSlices colorImage numTimePoints numSrcFiles firstOrigFile firstOrigFileWOPath firstOrigDir outFileName inputFilterLabel lossy compressionType humanReadableOrientation accessionNumber acquisitionDate acquisitionMatrix acquisitionNumber acquisitionTime aCR_NEMA_AcquisitionsInSeries aCR_NEMA_ImageOrientation aCR_NEMA_ImagePosition admittingDiagnosesDescription angioFlag bitsAllocated bitsStored bodyPartExamined codeMeaning codeValue columns contentDate contentTime contrastBolusAgent contrastBolusStartTime convolutionKernel currentPatientLocation derivationDescription echoTime echoTrainLength exposure exposureTime flipAngle frameOfReferenceUID gantryAngle gantryAngleTolerance gantryDetectorSlew gantryDetectorTilt gantryRotationDirection highBit imageComments imagedNucleus imageOrientationPatient imagePositionPatient imageType imagingFrequency instanceCreationDate instanceCreationTime instanceNumber institutionAddress institutionName inversionTime kVP largestImagePixelValue magneticFieldStrength manufacturer manufacturersModelName mediaStorageSOPInstanceUID modality mRAcquisitionType nameOfPhysiciansReadingStudy numberOfAverages numberOfFrames numberOfPhaseEncodingSteps numberOfTemporalPositions otherPatientNames patientComments patientID patientOrientation patientsAge percentSampling patientSex patientPosition patientsAddress patientsMothersBirthName patientsName patientsTelephoneNumbers patientsWeight patientsBirthDate performedProcedureStepDescription performedProcedureStepStartDate performedProcedureStepStartTime photometricInterpretation physiciansOfRecord pixelRepresentation pixelSpacing protocolName receiveCoilName reconstructionDiameter referringPhysiciansName repetitionTime requestedProcedureDescription requestingPhysician requestingService rescaleIntercept rescaleSlope rescaleType rows samplesPerPixel sAR scanningSequence scanOptions scheduledProcedureStepDescription sequenceName sequenceVariant seriesDate seriesDescription seriesInstanceUID seriesNumber seriesTime sliceLocation sliceThickness smallestImagePixelValue softwareVersions sOPClassUID sOPInstanceUID spacingBetweenSlices stationName studyDate studyDescription studyID studyInstanceUID studyTime windowCenter windowWidth xRayTubeCurrent¶
Defines the order and visibility of columns. All space separated entries in lines which do not start with ‘#’ will be interpreted as column codes. The first column in the Volume list will always be the volume index which shows the volume index according to
Sort Modeas a column in the volume list. The second column will be - if enabled - the volume thumbnail. All other columns can be configured in lines which do not start with a ‘#’.The following values are calculated:
validImage: Shows whether the volume is a valid image or not as a column in the volume list.
hasIssues: Shows whether the volume had warnings or errors while it was imported.
importTime: Time needed to compose this volume from already loaded DICOM files.
fileType: Shows the file format of the imported file as a column in the volume list, original:** if it is composed of multiple files:** the file format of the first file.
voxelType: Shows the voxel data type of the volume as a column in the volume list.
imgExt: Shows the extent of the volume as a column in the volume list.
voxelExtent: Shows the extent of voxels of the volume in millimeters as a column in the volume list.
numVoxels: Shows the number of voxels of the volume as a column in the volume list.
numSlices: Shows the number of slices of the volume as a column in the volume list.
colorImage: Shows whether the volume is a color image or not as a column in the volume list.
numTimePoints: Shows the number of time points of the volume as a column in the volume list.
numSrcFiles: Shows the number of files used to compose the volume as a column in the volume list.
firstOrigFile: Shows the path of the first file used to compose the volume as a column in the volume list.
outFileName: Shows the path to the created output file(s) without suffix (if created) as a column in the volume list.
inputFilterLabel: If enabled then a label assigned to any volume by an input filter will be shown as a column in the volume list view.
lossy: Shows the column in the volume list which indicates whether a volume is lossy compressed.
compressionType: Shows the column in the volume list which shows the compression type used to for this volume.
All other values represent the values of DICOM tags with corresponding names.
Min/Max Calculation¶
- name: dplCalcMinMax, type: Enum, default: Automatic, deprecated name: calcMinMax¶
Determines in which way the minimum and maximum voxel values are determined for imported volumes.
Values:
Title |
Name |
Description |
|---|---|---|
Automatic |
Automatic |
Used DICOM tags if available and scans image data for exact values if DICOM tags are not found. |
Only From Dicom Tags |
OnlyFromDicomTags |
Uses only DICOM tag values and sets min/max to data type range if tags are not found. |
Scan Image Data |
ScanImageData |
Always scans image data for exact values. This is the most reliable but also the most expensive mode. |
Anonymize name and birth date¶
- name: dplAnon, type: Bool, default: FALSE¶
This flag is provided only for backward compatibility, do not use! Its activation anonymizes patient id name and birth date on top level tags in composed structured multi-frame DICOM headers, but it affects no tags in multi-frame (sub)trees, nor in private tags nor in non-composed DICOM trees which are directly imported form single files. Therefore it is not sufficient to fulfill any typical anonymization requirements.
Verbose Mode¶
- name: dplVerbose, type: Enum, default: 0¶
Controls how much information is logged about volume sorting and splitting during import operations.
Values:
Title |
Name |
Description |
|---|---|---|
0 |
0 |
Only warnings and errors are shown. |
1 |
1 |
Warnings, errors, and further runtime information is shown. |
Special Processors¶
- name: specialProcessors, type: Enum, default: Off¶
Provides dedicated modes to (post)process imported volumes. Note that volume post-processing can degrade performance.
Values:
Title |
Name |
Description |
|---|---|---|
Off |
Off |
No postprocessing is performed. |
Process Set NGo Series |
ProcessSetNGoSeries |
Warning Experimental option: This post-processor tries to compose so-called Siemens Set’n’Go-Series to a valid volume by detecting best stitching positions and by removing overlapping frames (if there are any). Important
Note When importing with Set’N’Go post-processing you should expect that composed volumes have certain irregularities which are denoted with the following error and/or warning messages:
|
Process Set NGo Series Only |
ProcessSetNGoSeriesOnly |
Warning Experimental option: Only valid overlapping Siemens Set’n’Go-Series are processed, all others are discarded. This simplifies finding out which volumes are Set’n’Go ones. For details see ProcessSetNGoSeries. |
Minimum Num Frames in Volume¶
- name: minimumNumFramesInVolume, type: Integer, default: 1¶
Sets the minimum number of frames required in a sub-partition to break up an inhomogeneous partition into smaller potentially homogeneous partitions. If an inhomogeneous partition is broken up into smaller partitions, at least one of those partitions must have a size greater-than-or-equal-to this value.
In other words this parameter controls the activation of the irregular volume composition. Normally series with irregular slice distances are not composed but decomposed to a set of volumes where each volume obeys the given relative and absolute position and distance tolerances. If, however, the number of frames in each created volume is smaller than
Minimum Num Frames in Volumethen the irregular volume composition becomes active and builds a volume with irregular frames distances anyhow.
Dpl Config String0¶
- name: dplConfigString0, type: String, default: (, {Element = (0008,0060); Name = Modality; Sort = 1; Part = 1; },, {Element = (0008,0020); Name = StudyDate; Sort = 1; Part = 1; },, {Element = (0008,0008); Name = ImageType; Sort = 1; Part = 1; },, {Element = (0018,1030); Name = ProtocolName; Sort = 1; Part = 1; },, {Element = (0018,0050); Name = SliceThickness; Sort = 1; Part = 1; Tolerance = 0.000005; },, {Element = (0018,0080); Name = RepetitionTime; Sort = 1; Part = 1; },, {Element = (0018,0082); Name = InversionTime; Sort = 1; Part = 1; },, {Element = (0018,0091); Name = EchoTrainLength; Sort = 1; Part = 1; },, {Element = (0018,1210); Name = ConvolutionKernel; Sort = 1; Part = 1; },, {Element = (0018,1314); Name = FlipAngle; Sort = 1; Part = 1; },, {Element = (0018,0015); Name = BodyPartExamined; Sort = 1; Part = 1; },, {Element = (0028,0008); Name = NumberOfFrames; Sort = 1; Part = 1; },, {Element = (0028,0010); Name = Rows; Sort = 1; Part = 1; },, {Element = (0028,0011); Name = Columns; Sort = 1; Part = 1; },, {Element = (0020,0037); Name = ImageOrientationPatient; Sort = 1; Part = 1; Tolerance = 0.000005; },, {Element = (0018,1004); Name = PlateID; Sort = 1; Part = 1; },, {Element = (0018,1000); Name = DeviceSerialNumber; Sort = 1; Part = 1; },, {Element = (0028,0004); Name = PhotometricInterpretation; Sort = 1; Part = 1; },, {Element = (0028,0100); Name = BitsAllocated; Sort = 1; Part = 1; },, {Element = (0028,0102); Name = HighBit; Sort = 1; Part = 1; },, {Element = (0028,0103); Name = PixelRepresentation; Sort = 1; Part = 1; },, {Element = (0018,0020); Name = ScanningSequence; Sort = 1; Part = 1; },, {Element = (0018,0023); Name = MRAcquisitionType; Sort = 1; Part = 1; },, {Element = (0028,0030); Name = PixelSpacing; Sort = 1; Part = 1; Tolerance = 0.00001; },, {Element = (0054,0010); Name = EnergyWindowVector; Sort = 1; Part = 1; },, {Element = (0054,0030); Name = PhaseVector; Sort = 1; Part = 1; },, {Element = (0054,0060); Name = RRIntervalVector; Sort = 1; Part = 1; },, {Element = (0054,0050); Name = RotationVector; Sort = 1; Part = 1; },, {Element = (0028,1101); Name = RedPaletteColorLookupTableDescriptor; Sort = 1; Part = 1; },, {Element = (0028,1102); Name = GreenPaletteColorLookupTableDescriptor; Sort = 1; Part = 1; },, {Element = (0028,1103); Name = BluePaletteColorLookupTableDescriptor; Sort = 1; Part = 1; },, {Element = (0028,1201); Name = RedPaletteColorLookupTableData; Sort = 1; Part = 1; },, {Element = (0028,1202); Name = GreenPaletteColorLookupTableData; Sort = 1; Part = 1; },, {Element = (0028,1203); Name = BluePaletteColorLookupTableData; Sort = 1; Part = 1; },, {Element = (0020,000e); Name = SeriesInstanceUID; Sort = 1; SortCondition = "Modality = NM & !(FrameIncrementPointer = '(0054,0010)')" Part = 1; PartCondition = "Modality = NM & !(FrameIncrementPointer = '(0054,0010)')"; },, {Element = (0008,0018); Name = SOPInstanceUID; Sort = 1; SortCondition = "Modality = NM & NumberOfFrames > 1"; Part = 1; PartCondition = "Modality = NM & NumberOfFrames > 1"; },, {Element = (0008,0018); Name = SOPInstanceUID; Sort = 1; SortCondition = "Modality = AU | Modality = CR | Modality = DOC | Modality = DR | Modality = ECG | Modality = FID | Modality = KO | Modality = MG | Modality = MX | Modality = OCT | Modality = OT | Modality = PR | Modality = REG | Modality = RG | Modality = RTDOSE | Modality = RTIMAGE | Modality = RTPLAN | Modality = RTRAD | Modality = RTRECORD | Modality = RTSTRUCT | Modality = SEG | Modality = SR"; Part = 1; PartCondition = "Modality = AU | Modality = CR | Modality = DOC | Modality = DR | Modality = ECG | Modality = FID | Modality = KO | Modality = MG | Modality = MX | Modality = OCT | Modality = OT | Modality = PR | Modality = REG | Modality = RG | Modality = RTDOSE | Modality = RTIMAGE | Modality = RTPLAN | Modality = RTRAD | Modality = RTRECORD | Modality = RTSTRUCT | Modality = SEG | Modality = SR"; },, {Element = (0008,1090); Name = ManufacturerModelName; Sort = 1; Part = 1; },, {Element = (0008,1010); Name = StationName; Sort = 1; Part = 1; },, {Element = (0008,0070); Name = Manufacturer; Sort = 1; Part = 1; },, {Element = (0018,0020); Name = ScanningSequence; Sort = 1; Part = 1; },, {Element = (0018,0085); Name = ImagedNucleus; Sort = 1; Part = 1; },, {Element = (0018,0087); Name = MagneticFieldStrength; Sort = 1; Part = 1; },, {Element = (0018,1020); Name = SoftwareVersion; Sort = 1; Part = 1; },, {Element = (0018,5100); Name = PatientPosition; Sort = 1; Part = 1; },, {Element = (0062,000b); Name = ReferencedSegmentNumber; Sort = 1; SortCondition = "SOPClassUID = 1.2.840.10008.5.1.4.1.1.66.4"; Part = 1; PartCondition = "SOPClassUID = 1.2.840.10008.5.1.4.1.1.66.4"; },, {Element = (0020,000e); Name = SeriesInstanceUID; Sort = 1; SortCondition = "SOPClassUID = 1.2.840.10008.5.1.4.1.1.66.4"; Part = 1; PartCondition = "SOPClassUID = 1.2.840.10008.5.1.4.1.1.66.4"; },, {Element = (0020,0013); Name = InstanceNumber; Sort = 1; Part = 0; },, {Element = (0008,0030); Name = StudyTime; Sort = 1; Part = 0; },, {Element = (0020,0011); Name = SeriesNumber; Sort = 1; Part = 0; },, {Element = (0008,0021); Name = SeriesDate; Sort = 1; Part = 0; },, {Element = (0008,0031); Name = SeriesTime; Sort = 1; Part = 0; },, {Element = (0018,0081); Name = EchoTime; Sort = 1; Part = 0; },, {Element = (0018,0024); Name = SequenceName; Sort = 1; Part = 0; },, {Element = (0020,0012); Name = AcquisitionNumber; Sort = 1; Part = 0; },, {Element = (0018,0022); Name = ScanOptions; Sort = 1; Part = 0; },, {Element = (0008,0022); Name = AcquisitionDate; Sort = 1; Part = 0; },, {Element = (0008,0032); Name = AcquisitionTime; Sort = 1; Part = 0; },, {Element = (0008,0023); Name = ContentDate; Sort = 1; Part = 0; },, {Element = (0008,0033); Name = ContentTime; Sort = 1; Part = 0; },, {Element = (0020,0032); Name = ImagePositionPatient; Sort = 1; Part = 0; },, {Element = (0054,0020); Name = DetectorVector; Sort = 1; Part = 0; },, {Element = (0054,0100); Name = TimeSliceVector; Sort = 1; Part = 0; },, {Element = (0054,0070); Name = TimeSlotVector; Sort = 1; Part = 0; },, {Element = (0054,0080); Name = SliceVector; Sort = 1; Part = 0; },, {Element = (0054,0090); Name = AngularViewVector; Sort = 1; Part = 0; },, {Element = (0020,9157); Name = DimensionIndexValues; Sort = 1; Part = 0; },, ),¶
The default configuration used by the DPL for composing/sorting DICOM frames to volumes; see
Selected Configurationfor details.It cannot be modified by the user to always have the standard configuration available.
Dpl Config String1¶
- name: dplConfigString1, type: String, default: (, {Element = (0008,0060); Name = Modality; Sort = 1; Part = 1; },, {Element = (0008,0020); Name = StudyDate; Sort = 1; Part = 1; },, {Element = (0008,0008); Name = ImageType; Sort = 1; Part = 1; },, {Element = (0018,1030); Name = ProtocolName; Sort = 1; Part = 1; },, {Element = (0018,0050); Name = SliceThickness; Sort = 1; Part = 1; Tolerance = 0.000005; },, {Element = (0018,0080); Name = RepetitionTime; Sort = 1; Part = 1; },, {Element = (0018,0082); Name = InversionTime; Sort = 1; Part = 1; },, {Element = (0018,0091); Name = EchoTrainLength; Sort = 1; Part = 1; },, {Element = (0018,1210); Name = ConvolutionKernel; Sort = 1; Part = 1; },, {Element = (0018,1314); Name = FlipAngle; Sort = 1; Part = 1; },, {Element = (0018,0015); Name = BodyPartExamined; Sort = 1; Part = 1; },, {Element = (0028,0008); Name = NumberOfFrames; Sort = 1; Part = 1; },, {Element = (0028,0010); Name = Rows; Sort = 1; Part = 1; },, {Element = (0028,0011); Name = Columns; Sort = 1; Part = 1; },, {Element = (0020,0037); Name = ImageOrientationPatient; Sort = 1; Part = 1; Tolerance = 0.000005; },, {Element = (0018,1004); Name = PlateID; Sort = 1; Part = 1; },, {Element = (0018,1000); Name = DeviceSerialNumber; Sort = 1; Part = 1; },, {Element = (0028,0004); Name = PhotometricInterpretation; Sort = 1; Part = 1; },, {Element = (0028,0100); Name = BitsAllocated; Sort = 1; Part = 1; },, {Element = (0028,0102); Name = HighBit; Sort = 1; Part = 1; },, {Element = (0028,0103); Name = PixelRepresentation; Sort = 1; Part = 1; },, {Element = (0018,0020); Name = ScanningSequence; Sort = 1; Part = 1; },, {Element = (0018,0023); Name = MRAcquisitionType; Sort = 1; Part = 1; },, {Element = (0028,0030); Name = PixelSpacing; Sort = 1; Part = 1; Tolerance = 0.00001; },, {Element = (0054,0010); Name = EnergyWindowVector; Sort = 1; Part = 1; },, {Element = (0054,0030); Name = PhaseVector; Sort = 1; Part = 1; },, {Element = (0054,0060); Name = RRIntervalVector; Sort = 1; Part = 1; },, {Element = (0054,0050); Name = RotationVector; Sort = 1; Part = 1; },, {Element = (0028,1101); Name = RedPaletteColorLookupTableDescriptor; Sort = 1; Part = 1; },, {Element = (0028,1102); Name = GreenPaletteColorLookupTableDescriptor; Sort = 1; Part = 1; },, {Element = (0028,1103); Name = BluePaletteColorLookupTableDescriptor; Sort = 1; Part = 1; },, {Element = (0028,1201); Name = RedPaletteColorLookupTableData; Sort = 1; Part = 1; },, {Element = (0028,1202); Name = GreenPaletteColorLookupTableData; Sort = 1; Part = 1; },, {Element = (0028,1203); Name = BluePaletteColorLookupTableData; Sort = 1; Part = 1; },, {Element = (0020,000e); Name = SeriesInstanceUID; Sort = 1; SortCondition = "Modality = NM & !(FrameIncrementPointer = '(0054,0010)')" Part = 1; PartCondition = "Modality = NM & !(FrameIncrementPointer = '(0054,0010)')"; },, {Element = (0008,0018); Name = SOPInstanceUID; Sort = 1; SortCondition = "Modality = NM & NumberOfFrames > 1"; Part = 1; PartCondition = "Modality = NM & NumberOfFrames > 1"; },, {Element = (0008,0018); Name = SOPInstanceUID; Sort = 1; SortCondition = "Modality = AU | Modality = CR | Modality = DOC | Modality = DR | Modality = ECG | Modality = FID | Modality = KO | Modality = MG | Modality = MX | Modality = OCT | Modality = OT | Modality = PR | Modality = REG | Modality = RG | Modality = RTDOSE | Modality = RTIMAGE | Modality = RTPLAN | Modality = RTRAD | Modality = RTRECORD | Modality = RTSTRUCT | Modality = SEG | Modality = SR"; Part = 1; PartCondition = "Modality = AU | Modality = CR | Modality = DOC | Modality = DR | Modality = ECG | Modality = FID | Modality = KO | Modality = MG | Modality = MX | Modality = OCT | Modality = OT | Modality = PR | Modality = REG | Modality = RG | Modality = RTDOSE | Modality = RTIMAGE | Modality = RTPLAN | Modality = RTRAD | Modality = RTRECORD | Modality = RTSTRUCT | Modality = SEG | Modality = SR"; },, {Element = (0008,1090); Name = ManufacturerModelName; Sort = 1; Part = 1; },, {Element = (0008,1010); Name = StationName; Sort = 1; Part = 1; },, {Element = (0008,0070); Name = Manufacturer; Sort = 1; Part = 1; },, {Element = (0018,0020); Name = ScanningSequence; Sort = 1; Part = 1; },, {Element = (0018,0085); Name = ImagedNucleus; Sort = 1; Part = 1; },, {Element = (0018,0087); Name = MagneticFieldStrength; Sort = 1; Part = 1; },, {Element = (0018,1020); Name = SoftwareVersion; Sort = 1; Part = 1; },, {Element = (0018,5100); Name = PatientPosition; Sort = 1; Part = 1; },, {Element = (0062,000b); Name = ReferencedSegmentNumber; Sort = 1; SortCondition = "SOPClassUID = 1.2.840.10008.5.1.4.1.1.66.4"; Part = 1; PartCondition = "SOPClassUID = 1.2.840.10008.5.1.4.1.1.66.4"; },, {Element = (0020,000e); Name = SeriesInstanceUID; Sort = 1; SortCondition = "SOPClassUID = 1.2.840.10008.5.1.4.1.1.66.4"; Part = 1; PartCondition = "SOPClassUID = 1.2.840.10008.5.1.4.1.1.66.4"; },, {Element = (0020,0013); Name = InstanceNumber; Sort = 1; Part = 0; },, {Element = (0008,0030); Name = StudyTime; Sort = 1; Part = 0; },, {Element = (0020,0011); Name = SeriesNumber; Sort = 1; Part = 0; },, {Element = (0008,0021); Name = SeriesDate; Sort = 1; Part = 0; },, {Element = (0008,0031); Name = SeriesTime; Sort = 1; Part = 0; },, {Element = (0018,0081); Name = EchoTime; Sort = 1; Part = 0; },, {Element = (0018,0024); Name = SequenceName; Sort = 1; Part = 0; },, {Element = (0020,0012); Name = AcquisitionNumber; Sort = 1; Part = 0; },, {Element = (0018,0022); Name = ScanOptions; Sort = 1; Part = 0; },, {Element = (0008,0022); Name = AcquisitionDate; Sort = 1; Part = 0; },, {Element = (0008,0032); Name = AcquisitionTime; Sort = 1; Part = 0; },, {Element = (0008,0023); Name = ContentDate; Sort = 1; Part = 0; },, {Element = (0008,0033); Name = ContentTime; Sort = 1; Part = 0; },, {Element = (0020,0032); Name = ImagePositionPatient; Sort = 1; Part = 0; },, {Element = (0054,0020); Name = DetectorVector; Sort = 1; Part = 0; },, {Element = (0054,0100); Name = TimeSliceVector; Sort = 1; Part = 0; },, {Element = (0054,0070); Name = TimeSlotVector; Sort = 1; Part = 0; },, {Element = (0054,0080); Name = SliceVector; Sort = 1; Part = 0; },, {Element = (0054,0090); Name = AngularViewVector; Sort = 1; Part = 0; },, {Element = (0020,9157); Name = DimensionIndexValues; Sort = 1; Part = 0; },, ),¶
The first user configuration used by the DPL for composing/sorting DICOM frames to volumes; see
Selected Configurationfor examples and limitations.
Dpl Config String2¶
- name: dplConfigString2, type: String, default: (, {Element = (0008,0060); Name = Modality; Sort = 1; Part = 1; },, {Element = (0008,0020); Name = StudyDate; Sort = 1; Part = 1; },, {Element = (0008,0008); Name = ImageType; Sort = 1; Part = 1; },, {Element = (0018,1030); Name = ProtocolName; Sort = 1; Part = 1; },, {Element = (0018,0050); Name = SliceThickness; Sort = 1; Part = 1; Tolerance = 0.000005; },, {Element = (0018,0080); Name = RepetitionTime; Sort = 1; Part = 1; },, {Element = (0018,0082); Name = InversionTime; Sort = 1; Part = 1; },, {Element = (0018,0091); Name = EchoTrainLength; Sort = 1; Part = 1; },, {Element = (0018,1210); Name = ConvolutionKernel; Sort = 1; Part = 1; },, {Element = (0018,1314); Name = FlipAngle; Sort = 1; Part = 1; },, {Element = (0018,0015); Name = BodyPartExamined; Sort = 1; Part = 1; },, {Element = (0028,0008); Name = NumberOfFrames; Sort = 1; Part = 1; },, {Element = (0028,0010); Name = Rows; Sort = 1; Part = 1; },, {Element = (0028,0011); Name = Columns; Sort = 1; Part = 1; },, {Element = (0020,0037); Name = ImageOrientationPatient; Sort = 1; Part = 1; Tolerance = 0.000005; },, {Element = (0018,1004); Name = PlateID; Sort = 1; Part = 1; },, {Element = (0018,1000); Name = DeviceSerialNumber; Sort = 1; Part = 1; },, {Element = (0028,0004); Name = PhotometricInterpretation; Sort = 1; Part = 1; },, {Element = (0028,0100); Name = BitsAllocated; Sort = 1; Part = 1; },, {Element = (0028,0102); Name = HighBit; Sort = 1; Part = 1; },, {Element = (0028,0103); Name = PixelRepresentation; Sort = 1; Part = 1; },, {Element = (0018,0020); Name = ScanningSequence; Sort = 1; Part = 1; },, {Element = (0018,0023); Name = MRAcquisitionType; Sort = 1; Part = 1; },, {Element = (0028,0030); Name = PixelSpacing; Sort = 1; Part = 1; Tolerance = 0.00001; },, {Element = (0054,0010); Name = EnergyWindowVector; Sort = 1; Part = 1; },, {Element = (0054,0030); Name = PhaseVector; Sort = 1; Part = 1; },, {Element = (0054,0060); Name = RRIntervalVector; Sort = 1; Part = 1; },, {Element = (0054,0050); Name = RotationVector; Sort = 1; Part = 1; },, {Element = (0028,1101); Name = RedPaletteColorLookupTableDescriptor; Sort = 1; Part = 1; },, {Element = (0028,1102); Name = GreenPaletteColorLookupTableDescriptor; Sort = 1; Part = 1; },, {Element = (0028,1103); Name = BluePaletteColorLookupTableDescriptor; Sort = 1; Part = 1; },, {Element = (0028,1201); Name = RedPaletteColorLookupTableData; Sort = 1; Part = 1; },, {Element = (0028,1202); Name = GreenPaletteColorLookupTableData; Sort = 1; Part = 1; },, {Element = (0028,1203); Name = BluePaletteColorLookupTableData; Sort = 1; Part = 1; },, {Element = (0020,000e); Name = SeriesInstanceUID; Sort = 1; SortCondition = "Modality = NM & !(FrameIncrementPointer = '(0054,0010)')" Part = 1; PartCondition = "Modality = NM & !(FrameIncrementPointer = '(0054,0010)')"; },, {Element = (0008,0018); Name = SOPInstanceUID; Sort = 1; SortCondition = "Modality = NM & NumberOfFrames > 1"; Part = 1; PartCondition = "Modality = NM & NumberOfFrames > 1"; },, {Element = (0008,0018); Name = SOPInstanceUID; Sort = 1; SortCondition = "Modality = AU | Modality = CR | Modality = DOC | Modality = DR | Modality = ECG | Modality = FID | Modality = KO | Modality = MG | Modality = MX | Modality = OCT | Modality = OT | Modality = PR | Modality = REG | Modality = RG | Modality = RTDOSE | Modality = RTIMAGE | Modality = RTPLAN | Modality = RTRAD | Modality = RTRECORD | Modality = RTSTRUCT | Modality = SEG | Modality = SR"; Part = 1; PartCondition = "Modality = AU | Modality = CR | Modality = DOC | Modality = DR | Modality = ECG | Modality = FID | Modality = KO | Modality = MG | Modality = MX | Modality = OCT | Modality = OT | Modality = PR | Modality = REG | Modality = RG | Modality = RTDOSE | Modality = RTIMAGE | Modality = RTPLAN | Modality = RTRAD | Modality = RTRECORD | Modality = RTSTRUCT | Modality = SEG | Modality = SR"; },, {Element = (0008,1090); Name = ManufacturerModelName; Sort = 1; Part = 1; },, {Element = (0008,1010); Name = StationName; Sort = 1; Part = 1; },, {Element = (0008,0070); Name = Manufacturer; Sort = 1; Part = 1; },, {Element = (0018,0020); Name = ScanningSequence; Sort = 1; Part = 1; },, {Element = (0018,0085); Name = ImagedNucleus; Sort = 1; Part = 1; },, {Element = (0018,0087); Name = MagneticFieldStrength; Sort = 1; Part = 1; },, {Element = (0018,1020); Name = SoftwareVersion; Sort = 1; Part = 1; },, {Element = (0018,5100); Name = PatientPosition; Sort = 1; Part = 1; },, {Element = (0062,000b); Name = ReferencedSegmentNumber; Sort = 1; SortCondition = "SOPClassUID = 1.2.840.10008.5.1.4.1.1.66.4"; Part = 1; PartCondition = "SOPClassUID = 1.2.840.10008.5.1.4.1.1.66.4"; },, {Element = (0020,000e); Name = SeriesInstanceUID; Sort = 1; SortCondition = "SOPClassUID = 1.2.840.10008.5.1.4.1.1.66.4"; Part = 1; PartCondition = "SOPClassUID = 1.2.840.10008.5.1.4.1.1.66.4"; },, {Element = (0020,0013); Name = InstanceNumber; Sort = 1; Part = 0; },, {Element = (0008,0030); Name = StudyTime; Sort = 1; Part = 0; },, {Element = (0020,0011); Name = SeriesNumber; Sort = 1; Part = 0; },, {Element = (0008,0021); Name = SeriesDate; Sort = 1; Part = 0; },, {Element = (0008,0031); Name = SeriesTime; Sort = 1; Part = 0; },, {Element = (0018,0081); Name = EchoTime; Sort = 1; Part = 0; },, {Element = (0018,0024); Name = SequenceName; Sort = 1; Part = 0; },, {Element = (0020,0012); Name = AcquisitionNumber; Sort = 1; Part = 0; },, {Element = (0018,0022); Name = ScanOptions; Sort = 1; Part = 0; },, {Element = (0008,0022); Name = AcquisitionDate; Sort = 1; Part = 0; },, {Element = (0008,0032); Name = AcquisitionTime; Sort = 1; Part = 0; },, {Element = (0008,0023); Name = ContentDate; Sort = 1; Part = 0; },, {Element = (0008,0033); Name = ContentTime; Sort = 1; Part = 0; },, {Element = (0020,0032); Name = ImagePositionPatient; Sort = 1; Part = 0; },, {Element = (0054,0020); Name = DetectorVector; Sort = 1; Part = 0; },, {Element = (0054,0100); Name = TimeSliceVector; Sort = 1; Part = 0; },, {Element = (0054,0070); Name = TimeSlotVector; Sort = 1; Part = 0; },, {Element = (0054,0080); Name = SliceVector; Sort = 1; Part = 0; },, {Element = (0054,0090); Name = AngularViewVector; Sort = 1; Part = 0; },, {Element = (0020,9157); Name = DimensionIndexValues; Sort = 1; Part = 0; },, ),¶
The second user configuration used by the DPL for composing/sorting DICOM frames to volumes; see
Selected Configurationfor examples and limitations.
Dpl Config String3¶
- name: dplConfigString3, type: String, default: (, {Element = (0008,0060); Name = Modality; Sort = 1; Part = 1; },, {Element = (0008,0020); Name = StudyDate; Sort = 1; Part = 1; },, {Element = (0008,0008); Name = ImageType; Sort = 1; Part = 1; },, {Element = (0018,1030); Name = ProtocolName; Sort = 1; Part = 1; },, {Element = (0018,0050); Name = SliceThickness; Sort = 1; Part = 1; Tolerance = 0.000005; },, {Element = (0018,0080); Name = RepetitionTime; Sort = 1; Part = 1; },, {Element = (0018,0082); Name = InversionTime; Sort = 1; Part = 1; },, {Element = (0018,0091); Name = EchoTrainLength; Sort = 1; Part = 1; },, {Element = (0018,1210); Name = ConvolutionKernel; Sort = 1; Part = 1; },, {Element = (0018,1314); Name = FlipAngle; Sort = 1; Part = 1; },, {Element = (0018,0015); Name = BodyPartExamined; Sort = 1; Part = 1; },, {Element = (0028,0008); Name = NumberOfFrames; Sort = 1; Part = 1; },, {Element = (0028,0010); Name = Rows; Sort = 1; Part = 1; },, {Element = (0028,0011); Name = Columns; Sort = 1; Part = 1; },, {Element = (0020,0037); Name = ImageOrientationPatient; Sort = 1; Part = 1; Tolerance = 0.000005; },, {Element = (0018,1004); Name = PlateID; Sort = 1; Part = 1; },, {Element = (0018,1000); Name = DeviceSerialNumber; Sort = 1; Part = 1; },, {Element = (0028,0004); Name = PhotometricInterpretation; Sort = 1; Part = 1; },, {Element = (0028,0100); Name = BitsAllocated; Sort = 1; Part = 1; },, {Element = (0028,0102); Name = HighBit; Sort = 1; Part = 1; },, {Element = (0028,0103); Name = PixelRepresentation; Sort = 1; Part = 1; },, {Element = (0018,0020); Name = ScanningSequence; Sort = 1; Part = 1; },, {Element = (0018,0023); Name = MRAcquisitionType; Sort = 1; Part = 1; },, {Element = (0028,0030); Name = PixelSpacing; Sort = 1; Part = 1; Tolerance = 0.00001; },, {Element = (0054,0010); Name = EnergyWindowVector; Sort = 1; Part = 1; },, {Element = (0054,0030); Name = PhaseVector; Sort = 1; Part = 1; },, {Element = (0054,0060); Name = RRIntervalVector; Sort = 1; Part = 1; },, {Element = (0054,0050); Name = RotationVector; Sort = 1; Part = 1; },, {Element = (0028,1101); Name = RedPaletteColorLookupTableDescriptor; Sort = 1; Part = 1; },, {Element = (0028,1102); Name = GreenPaletteColorLookupTableDescriptor; Sort = 1; Part = 1; },, {Element = (0028,1103); Name = BluePaletteColorLookupTableDescriptor; Sort = 1; Part = 1; },, {Element = (0028,1201); Name = RedPaletteColorLookupTableData; Sort = 1; Part = 1; },, {Element = (0028,1202); Name = GreenPaletteColorLookupTableData; Sort = 1; Part = 1; },, {Element = (0028,1203); Name = BluePaletteColorLookupTableData; Sort = 1; Part = 1; },, {Element = (0020,000e); Name = SeriesInstanceUID; Sort = 1; SortCondition = "Modality = NM & !(FrameIncrementPointer = '(0054,0010)')" Part = 1; PartCondition = "Modality = NM & !(FrameIncrementPointer = '(0054,0010)')"; },, {Element = (0008,0018); Name = SOPInstanceUID; Sort = 1; SortCondition = "Modality = NM & NumberOfFrames > 1"; Part = 1; PartCondition = "Modality = NM & NumberOfFrames > 1"; },, {Element = (0008,0018); Name = SOPInstanceUID; Sort = 1; SortCondition = "Modality = AU | Modality = CR | Modality = DOC | Modality = DR | Modality = ECG | Modality = FID | Modality = KO | Modality = MG | Modality = MX | Modality = OCT | Modality = OT | Modality = PR | Modality = REG | Modality = RG | Modality = RTDOSE | Modality = RTIMAGE | Modality = RTPLAN | Modality = RTRAD | Modality = RTRECORD | Modality = RTSTRUCT | Modality = SEG | Modality = SR"; Part = 1; PartCondition = "Modality = AU | Modality = CR | Modality = DOC | Modality = DR | Modality = ECG | Modality = FID | Modality = KO | Modality = MG | Modality = MX | Modality = OCT | Modality = OT | Modality = PR | Modality = REG | Modality = RG | Modality = RTDOSE | Modality = RTIMAGE | Modality = RTPLAN | Modality = RTRAD | Modality = RTRECORD | Modality = RTSTRUCT | Modality = SEG | Modality = SR"; },, {Element = (0008,1090); Name = ManufacturerModelName; Sort = 1; Part = 1; },, {Element = (0008,1010); Name = StationName; Sort = 1; Part = 1; },, {Element = (0008,0070); Name = Manufacturer; Sort = 1; Part = 1; },, {Element = (0018,0020); Name = ScanningSequence; Sort = 1; Part = 1; },, {Element = (0018,0085); Name = ImagedNucleus; Sort = 1; Part = 1; },, {Element = (0018,0087); Name = MagneticFieldStrength; Sort = 1; Part = 1; },, {Element = (0018,1020); Name = SoftwareVersion; Sort = 1; Part = 1; },, {Element = (0018,5100); Name = PatientPosition; Sort = 1; Part = 1; },, {Element = (0062,000b); Name = ReferencedSegmentNumber; Sort = 1; SortCondition = "SOPClassUID = 1.2.840.10008.5.1.4.1.1.66.4"; Part = 1; PartCondition = "SOPClassUID = 1.2.840.10008.5.1.4.1.1.66.4"; },, {Element = (0020,000e); Name = SeriesInstanceUID; Sort = 1; SortCondition = "SOPClassUID = 1.2.840.10008.5.1.4.1.1.66.4"; Part = 1; PartCondition = "SOPClassUID = 1.2.840.10008.5.1.4.1.1.66.4"; },, {Element = (0020,0013); Name = InstanceNumber; Sort = 1; Part = 0; },, {Element = (0008,0030); Name = StudyTime; Sort = 1; Part = 0; },, {Element = (0020,0011); Name = SeriesNumber; Sort = 1; Part = 0; },, {Element = (0008,0021); Name = SeriesDate; Sort = 1; Part = 0; },, {Element = (0008,0031); Name = SeriesTime; Sort = 1; Part = 0; },, {Element = (0018,0081); Name = EchoTime; Sort = 1; Part = 0; },, {Element = (0018,0024); Name = SequenceName; Sort = 1; Part = 0; },, {Element = (0020,0012); Name = AcquisitionNumber; Sort = 1; Part = 0; },, {Element = (0018,0022); Name = ScanOptions; Sort = 1; Part = 0; },, {Element = (0008,0022); Name = AcquisitionDate; Sort = 1; Part = 0; },, {Element = (0008,0032); Name = AcquisitionTime; Sort = 1; Part = 0; },, {Element = (0008,0023); Name = ContentDate; Sort = 1; Part = 0; },, {Element = (0008,0033); Name = ContentTime; Sort = 1; Part = 0; },, {Element = (0020,0032); Name = ImagePositionPatient; Sort = 1; Part = 0; },, {Element = (0054,0020); Name = DetectorVector; Sort = 1; Part = 0; },, {Element = (0054,0100); Name = TimeSliceVector; Sort = 1; Part = 0; },, {Element = (0054,0070); Name = TimeSlotVector; Sort = 1; Part = 0; },, {Element = (0054,0080); Name = SliceVector; Sort = 1; Part = 0; },, {Element = (0054,0090); Name = AngularViewVector; Sort = 1; Part = 0; },, {Element = (0020,9157); Name = DimensionIndexValues; Sort = 1; Part = 0; },, ),¶
The third user configuration used by the DPL for composing/sorting DICOM frames to volumes; see
Selected Configurationfor examples and limitations.
Selected Configuration¶
- name: dplUsedConfig, type: Enum, default: DefaultConfig¶
Selects which DPL configuration shall be used for importing.
Notes:
Configuration lines containing Part = 1 should be listed first, then those with Part = 1 with conditions, and finally those with Part = 0.
The configuration selected by DefaultConfig cannot be modified, however, it is stored with the module state. So it always maintains the configuration state from the creation time of the module.
Be sure to have spaces between a description key, the “=”, and its value, so, for example, use Sort = 1 instead of Sort=1.
If tags have different tag values in different DICOM frames then they can be used for sorting or partitioning if they are listed in the used configuration string. In other words: If tags do not differ between frames, they normally will not be used for sorting or partitioning.
Tag descriptions in a configuration are ordered: If both tags, X and Y, have changing values between DICOM frames then tag X is used instead of Y for sorting or partitioning if it listed in the configuration before the tag Y.
Setting Sort = 0 also deactivates partitioning.
Conditions for sorting or partitioning can be added to the configuration, such as PartCondition = “<expr>” or SortCondition = “<expr>”. In expr the following operators can be used:
= for strings: case-insensitive check for equality, otherwise check for equality
== for strings: case-sensitive check for equality, otherwise the same as =
like (only for strings): case-insensitive substring check
<, <=, >=, >, != the usual arithmetic operators (do not use for strings)
Some examples:
- To avoid that different tag values between DICOM frames are used for partitioning set Part = 0:
{Element = (0028,0030); Name = PixelSpacing; Sort = 1; Part = 0; }
- To avoid that rounding errors smaller than 0.00001 in DICOM tags cause partitioning use Tolerance = <value>:
{Element = (0028,0030); Name = PixelSpacing; Sort = 1; Part = 1; Tolerance = 0.00001; }
- Apply parting only on specific modalities by using a PartCondition:
{Element = (0008,0018); Name = SOPInstanceUID; Sort = 1; Part = 1; PartCondition = “Modality = CR | Modality = DR | Modality = MG | Modality = MX”; }
- Use SortCondition to activate sorting (and therefore also partitioning) on SliceThickness if SeriesDescription does not contain the substring “mip”:
{Element = “(0018,0050)”; Name = SliceThickness; Part = 1; Sort = 1; SortCondition = “! SeriesDescription like mip”; }
Values:
Title |
Name |
|---|---|
Default Config |
DefaultConfig |
User Config1 |
UserConfig1 |
User Config2 |
UserConfig2 |
User Config3 |
UserConfig3 |
Relative Distance Tolerance¶
- name: relativeDistanceTolerance, type: Double, default: 0.25, minimum: 1e-07¶
The maximum difference between 2 slice distances in percents that does not split up a 3D volume (0 means 0%, 1 means 100%).
Absolute Distance Tolerance¶
- name: absoluteDistanceTolerance, type: Double, default: 0.02, minimum: 1e-07¶
The maximum difference between 2 slice distances in mm that does not split up a 3D volume.
Position Tolerance MM¶
- name: positionToleranceMM, type: Double, default: 0.0999¶
Absolute tolerance in mm, added to the previous value to decide if a volume is homogeneous.
Series Based Preprocessing Condition¶
- name: seriesBasedPreprocessingCondition, type: Bool, default: FALSE, deprecated name: setSeriesBasedPreprocessingCondition¶
If enabled then sorting and parting on DICOM frames with identical Series Instance UID values is changed. One of its effects is that the creation of volumes with multiple time points from such frames is suppressed in favor of possibly inhomogeneous 3D volume creation. Enable this with care and only on the smallest possibly subset of frames, for example by using a
FileListFilterPlugin.
Force2DPlus TCondition¶
- name: force2DPlusTCondition, type: String, default: Modality == XA | Modality == US | Modality == RF¶
A DICOM tag condition string; if evaluates to true for the first image in an image set, a 2D+T image set will be created (instead of a 3D or inhomogeneous image).
Log Parting And Sorting Information¶
- name: logPartingAndSortingInformation, type: Bool, default: FALSE¶
If enabled then partitioning and sorting information during the import process is logged independently from the verbose level in DPL imports.
Scan recursively¶
- name: dplScanRecursively, type: Bool, default: TRUE¶
If enabled then the during any importing step for all given directory paths the subdirectories will also be scanned recursively. If disabled then only flat scans of the given directories are done.
Decompose Multi Frame Files¶
- name: decomposeMultiFrameFiles, type: Bool, default: TRUE¶
If enabled then multiframe files are imported as bunches of single frames such that they can be sorted and partitioned correctly into different volumes, if necessary. WARNING: Although disabling this checkbox can improve import times significantly it also risks wrong frames orders, wrong volume subdivisions or wrong world matrices and voxel spacings. Disabling is only a backward compatibility option for some old importers which did not support enhanced multiframe files. Importing with this checkbox enabled is the strongly recommended option, because it leads to more correct results. This flag does not influence imports of single frame files or old-style multiframe files.
Copy Full Functional Group Sequences¶
- name: copyFullFunctionalGroupSequences, type: Bool, default: TRUE¶
When decomposing enhanced multi-frame files (see
Decompose Multi Frame Files) older versions ofDirectDicomImportdid not fully copy the structure of Functional Group Macros from source to decomposed DICOM frames. Now this has been added and usually defaults to true. However, older stored tests, networks or applications may not expect this additional information and are reloaded with disabled default until they are explicitly re-saved with an enabled one.
Remove private MFSQ tag¶
- name: removePrivateMFSQTag, type: Bool, default: FALSE¶
If true then the sequence tag with private multiframe tag information will be removed from created multiframe tree before appending it to the image properties, otherwise it will normally be appended. Note that this influences only the ML output image but not the internally maintained version of the tree, i.e. the shown shown tag dump in ‘Tags’ will still show the frame information.
Force Orthogonal World Matrix¶
- name: forceOrthogonalWorldMatrix, type: Bool, default: FALSE¶
If disabled (recommended) then the world matrix is determined from the ImageOrientationPatient vectors of the first frame for x/y and the distance vector between the two ImagePositionPatient positions of the first two frames. This is typically more reliable, especially in cases with gantry tilt scans. If enabled (not recommended) then the world matrix is always determined from the two ImageOrientationPatient vectors of the first frame in x/y and and their cross product for z. Warning: This is a backward compatibility mode, thus enable this only with care, for example in cases where ImagePositionPatient values for frames is not reliable.
Split time points to 3D volumes¶
- name: decomposeTo3DVolumes, type: Bool, default: FALSE¶
If enabled then volumes with a temporal extent are decomposed to multiple 3D volumes if these 3D volumes are build from different frame sets, otherwise, if false, volumes are composed in time dimension normally. Single file multiframe volumes having temporal extents are not affected.
Allow Mixed Pixel Types¶
- name: allowMixedPixelTypes, type: Bool, default: FALSE¶
Warning
Experimental option: It might not be stable for all configurations and therefore it should only be used on smallest possible subsets of data, for example by using a
FileListFilterPlugin. If enabled then the ML data type determined from the imported frame(s) is checked against the min/max values determined according to theMin/Max Calculationmode. If different pixel types in the imported frames appear then this data type might be too small to hold all values and must be enlarged. This flag only will have effect to the imported volumes if parting on data type relevant tags is disabled, which means that HighBit, BitsStored, BitsAllocated, and PixelRepresentation may not be used for parting in the DPL configuration which influences volume compositions. This mode is usually needed only in rare cases where DICOM files shall be composed which belong to the same series but which have different pixel representations or different BitsAllocated tags.
Disable DCMTree Normalization¶
- name: disableDCMTreeNormalization, type: Bool, default: FALSE¶
Warning
Experimental option: If true and multiple DICOM frames are imported, then the DCMTree is not normalized which may lead to significant larger structured multi-frame trees, but the tree structure is much more straight forward. Then each frame still contains nearly all of its original information, but the generated structure is non MeVisLab-standard and not necessarily supported in other modules any more; therefore the recommended default is off. It is not recommended to store such imported volumes or to make them persistent in any other way. The contents of the generated DCMTree structured are still subject to change. Note that you may also want to enable
Auto Create Missing Private Creatorsif you also want to see them in frames; this is not done automatically when this option is enabled.
Auto Create Missing Private Creators¶
- name: autoCreateMissingPrivateCreators, type: Bool, default: FALSE¶
Warning
Experimental option: Unfortunately in some incorrectly de-identified DICOM files, private creator tags have been removed which makes the depending private tags unusable. Also the creation of the structured multi-frame trees in the volume composition of
DirectDicomImportskips/removes such private tags which also makes them unaccessible in composed volumes. With enabledAuto Create Missing Private Creators, missing private creator tags are automatically created which makes the private tags remain. However, it is left to the user what to do with them, because without additional knowledge about the original private creator(s) they are usually useless. Therefore use this option with care, default is off.
Dpl Hard Overwrite Tag On0¶
- name: dplHardOverwriteTagOn0, type: Bool, default: FALSE¶
If enabled then all tags with given Id are overwritten with Value before using it in DPL sorting and parting process. The changed tag values are not used outside this process, thus they do not appear the created output volumes or tag dumps.
Id (dplHardOverwriteTagId0)¶
- name: dplHardOverwriteTagId0, type: String¶
Tag id in format (XYZC,ABCD) specifying which tag is overwritten with Value before feeding them into DPL sorting and parting process. DICOM tags in the created output volume are not affected.
Dpl Hard Overwrite Tag On1¶
- name: dplHardOverwriteTagOn1, type: Bool, default: FALSE¶
If enabled then all tags with given id are overwritten with Value before using it in DPL sorting and parting process. Tags not used in this process and in the created output volume are not affected, so this feature is limited.
Id (dplHardOverwriteTagId1)¶
- name: dplHardOverwriteTagId1, type: String¶
Another setting such as dplHardOverwriteTagId0.
Dpl Hard Overwrite Tag On2¶
- name: dplHardOverwriteTagOn2, type: Bool, default: FALSE¶
If enabled then all tags with given id are overwritten with Value before using it in DPL sorting and parting process. Tags not used in this process and in the created output volume are not affected, so this feature is limited.
Id (dplHardOverwriteTagId2)¶
- name: dplHardOverwriteTagId2, type: String¶
Another setting such as dplHardOverwriteTagId0.
Dpl Hard Overwrite Tag On3¶
- name: dplHardOverwriteTagOn3, type: Bool, default: FALSE¶
If enabled then all tags with given id are overwritten with Value before using it in DPL sorting and parting process. Tags not used in this process and in the created output volume are not affected, so this feature is limited.
Id (dplHardOverwriteTagId3)¶
- name: dplHardOverwriteTagId3, type: String¶
Another setting such as dplHardOverwriteTagId0.
Allow Sorting According to DimensionIndexValues Tag¶
- name: dplAllowSortingAccordingToDimensionIndexValues, type: Enum, default: Auto¶
Handle the special case where the ImagePositionPatient tag is legally missing on images, i.e. if the Plane Position (Patient) Macro is not required in the Per-Frame functional groups sequence, for example in multi-frame US data. Alternatively the sorting, e.g. on US modality images, sometimes can be performed using the Dimension Index Values tag instead. If this field is Auto then this sorting is active. Otherwise sorting will not be done geometrically. See also the DICOM standard, C.7.6.16 Multi-frame Functional Groups Module.
Values:
Title |
Name |
|---|---|
Auto |
Auto |
Off |
Off |
Replacement for Missing ImageOrientationPatient¶
- name: dplWorkaroundValueForImageOrientationPatientTag, type: String¶
If a DICOM frame has an ImagePositionPatient tag but no or an incomplete ImageOrientationPatient tag then this value will be used as replacement if it is not empty. This can be used to import partially corrupted data sets where ImageOrientationPatient is missing. A useful value might be 1\0\0\0\1\0.
Compose Other Files Mode¶
- name: composeOtherFilesMode, type: Enum, default: Single¶
Defines how same sized non DICOM files are added to the volume list.
Values:
Title |
Name |
Description |
|---|---|---|
Single |
Single |
Each file is imported as one single volumes. |
Heap |
Heap |
Files matching in extent, voxel type and color model are composed to a a heap in the next higher unused dimension. This is not performed with image data files which have DICOM information (such as DCM/Tiff pairs), because their DICOM information usually cannot be composed reliably to a new DICOM header. They will always be imported as single volume. |
Silent Trial File Types¶
- name: silentTrialFileTypes, type: String, default: .dat .raw .txt¶
If any files cannot be opened for any reason then this usually shall be logged for further analysis. In some cases, however, this is not desired: For example some files of type .raw can be loaded by plugins if they have appropriate format contents. Most other .raw files, however, are usually not automatically readable and should be ignored therefore. An error logging would not provide useful information. For this purpose in
Silent Trial File Typesa space separated list of file names and/or suffixes can be specified which shall be handled silently. Use this list with care to avoid that important error logging is suppressed!
Source Dir¶
- name: source, type: String¶
The directory (tree) to be parsed for DICOM files to be composed to volumes. It can be useful to leave this field empty, especially since
Input directory listandInput file listallow additional input sources. See also the buttons Add Source Directory… and Add Source Files… in Configuration -> Import Directories and Configuration -> Import Files.
Cache Path¶
- name: guiCachePath, type: String, deprecated name: cachePath¶
Directory where
DirectDicomImportcan store cache files to speed up file reloads etc. Note that this path is insensitive ifUse result cachingis disabled.