DicomImport¶
- MLModule¶
genre
author
package
dll
definition
see also
keywords
Purpose¶
The DicomImport module allows for importing DICOM files directly via a call to the Dicom Processor Library (DPL) without requiring an intermediate image representation.
After importing from a specified directory or a list of files, all volumes will be displayed in a tree structure containing patients, studies, and volumes.
The module output consists of images loaded from the DICOM frames. The images’ pixel data is only loaded when needed to reduce memory load.
Settings for volume creation and the DPL can be defined in the Configuration dialog. The DPL settings can be configured manually or by loading predefined files.
Usage¶
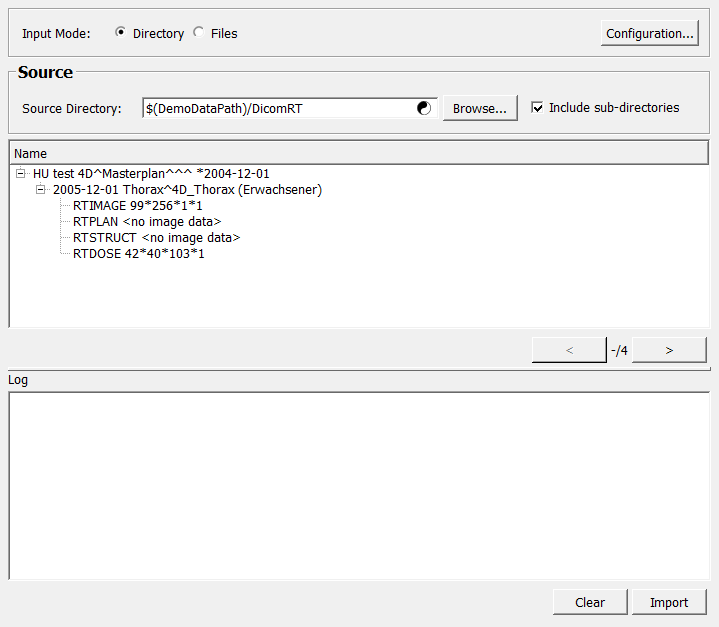
Choose between the two different input modes: from a directory or a list of files with the switch Input Mode.
Either specify the directory via the Source Directory or set a list of fully qualified file names Files.
Then use the Import button to parse the directories. This composes image volumes from the DICOM files in them and shows them in the patient tree.
Check the console and the outLogModel for issues, errors, and other information regarding the generated image volumes.
Select any composed volume by clicking on it. The selected volume is made available at output0.
Sorting and partitioning can be configured directly via Direct Sort Part Configuration or by selecting a file containing the configuration via Sort Part Config File.
In the event that multiple volumes have been generated during import, it is possible to toggle through the volumes forward (>) and backward (<). The total number of volumes generated can be found in the field volumeCount.
- Note:
After the import, the imported DICOM files are still needed for the image data in the image output, which will be read on demand. Under Windows, they will be locked and cannot be moved away until the import is cleared.
Details¶
Known Issues¶
- There are several reasons why importing can become slow or might not work; the most important and typical ones are listed here:
Float value pixel data files (for example, used for parametric maps) are currently not supported.
DICOM objects with an uncompressed transfer syntax and the stored Photometric Interpretations YBR_FULL_422, YBR_PARTIAL_422, and YBR_PARTIAL_420 are currently not displayed.
Interaction¶
You can expand and collapse the patient trees via a context menu on that area.
Tips¶
Composing volumes from DICOM source files depends on many factors, such as scan parameters and import settings.
DicomImportuses heuristics from the Dicom Processor Library (DPL). Try different parameter combinations if the volume composition is not as expected.When single or multiple DICOM frames are imported and composed into a volume, the page extent for the result volume is set to the extent of a single frame. This can lead to memory trouble if large frames are composed. In some cases, the usage of a
ImagePropertyConvertmodule afterDicomImportmay help to compensate for this problem.In the event of issues, you can define the logging level of the module via field
logLevel.
Windows¶
Default Panel¶
Configuration¶
FileList¶
Input Fields¶
inputFilterPlugin¶
- name: inputFilterPlugin, type: DicomImportFilterBase(MLBase)¶
Output Fields¶
output0¶
- name: output0, type: Image¶
Output connector providing image data of the currently selected volume.
outDicomTree¶
- name: outDicomTree, type: DicomTree(MLBase)¶
The DICOM tree of the currently selected item.
This is the same tree as provided with the image output, but is the only way to access the DICOM tree if the selected item does not contain image data.
For accessing this object via scripting, see the Scripting Reference:
MLABDicomTree.
outLogModel¶
- name: outLogModel, type: StandardItemModel(MLBase)¶
Output connector providing log information including a time stamp and error codes. Each entry corresponds to a warning or error during the import.
An error means that a file could not be processed. All errors are listed as top-level entries in the log model, with one of the error codes described below in the errorcode column.
A warning means that a file was processed but had some incorrect tags or other problems. All warnings related to a given input file are grouped under a single entry that contains the file path. Each warning has a warning code described below, which appears in the errorcode column.
- The following attributes are available:
timestamp - The time of the log is represented as the number of milliseconds elapsed since the epoch (January 1, 1970, 00:00:00)
path - The full path of the problematic input file; only given for errors and top-level warning entries
tag - If applicable, the tag causing the problem in the format “(gggg, eeee)”; otherwise, the tag is left empty
tagname - The tag name for “tag”, if it is known
errorcode - A numerical representation of the error or warning, see below
message - A textual representation of the error or warning
- The following error code are currently available:
1 - Unable to open the DICOM file
2 - The file does not seem to be a DICOM file
3 - The DICOM transfer syntax is not supported
4 - Mandatory DICOM tags needed for processing are missing
5 - Error while reading the DICOM file (errors from the underlying DICOM library)
6 - Warnings have occurred (for the top-level warning entry)
- The following warning codes are used:
1 - A value is incorrectly encoded
2 - The specific character set is invalid
3 - Warnings while reading the DICOM file (warnings from the underlying DICOM library)
4 - An incorrect Value Representation has been encountered
For accessing this object via scripting, see the Scripting Reference:
MLStandardItemModelWrapper.
outputModel¶
- name: outputModel, type: StandardItemModel(MLBase)¶
Output connector providing the patient tree(s) of the imported directory.
This lists the imported volumes in a hierarchy consisting of patients, studies, and imported volumes.
- This tree has two attributes: name and ID:
On the patient level, the name is the PatientName plus the PatientBirthDate (if given).
On the study level, the name is the StudyDate and StudyDescription.
On the volume level, the name is a combination of Modality, SeriesDescription (only in the case that it is the same for all included frames), and the size of the imported volume (“<Columns>*<Rows>*<slices>*<time points>”), or “<no image data>” if there was no image data for the item. The ID is the identification number of the imported item, unique across the entire current MeVisLab instance. This number can be used to select the item as output through the
selectedItemfield (alsoDicomImportExtraOutput.selectedItem), or as argument for theDicomImportResultWrapperwrapper ofoutImportResult.
For accessing this object via scripting, see the Scripting Reference:
MLStandardItemModelWrapper.
outImportResult¶
- name: outImportResult, type: DicomImportResult(MLBase)¶
This output field contains all import results. You can access this via scripting, or connect a
DicomImportExtraOutputmodule.For accessing this object via scripting, see the Scripting Reference:
DicomImportResultWrapper.
Parameter Fields¶
Field Index¶
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Visible Fields¶
Source Directory¶
- name: source, type: String¶
Sets the directory (tree) to be parsed for DICOM files.
Files¶
- name: files, type: String¶
Sets a list of strings that contain the fully qualified file names, each separated by a newline.
Input Mode¶
- name: inputMode, type: Enum, default: Directory¶
Defines the mode to control the input source.
Values:
Title |
Name |
Description |
|---|---|---|
Directory |
Directory |
If this is set, then the |
Files |
Files |
If this is set, then the |
Include sub-directories¶
- name: enableRecursiveSearch, type: Bool, default: TRUE¶
If checked, the given
Source Directoryis parsed including subdirectories.
Import¶
Clear¶
- name: clearImport, type: Trigger, deprecated name: stopImport¶
When pressed, the import of DICOM files is aborted if an import is currently in progress. Otherwise, it just clears the output.
Use file for sort/part¶
- name: useSortPartConfigFile, type: Bool, default: FALSE¶
If checked, the sort/part configuration is loaded from file given in
Sort Part Config File; otherwise, it is taken fromDirect Sort Part Configuration.
Sort Part Config File¶
- name: sortPartConfigFile, type: String¶
Sets the full path to the file that provides the sort/part configuration.
Used if
Use file for sort/partis True.
Direct Sort Part Configuration¶
- name: directSortPartConfiguration, type: String, default: (, {Element = (0008,0060); Name = Modality; Sort = 1; Part = 1; },, {Element = (0008,0020); Name = StudyDate; Sort = 1; Part = 1; },, {Element = (0008,0008); Name = ImageType; Sort = 1; Part = 1; },, {Element = (0018,1030); Name = ProtocolName; Sort = 1; Part = 1; },, {Element = (0018,0050); Name = SliceThickness; Sort = 1; Part = 1; Tolerance = 0.000005; },, {Element = (0018,0080); Name = RepetitionTime; Sort = 1; Part = 1; },, {Element = (0018,0082); Name = InversionTime; Sort = 1; Part = 1; },, {Element = (0018,0091); Name = EchoTrainLength; Sort = 1; Part = 1; },, {Element = (0018,1210); Name = ConvolutionKernel; Sort = 1; Part = 1; },, {Element = (0018,1314); Name = FlipAngle; Sort = 1; Part = 1; },, {Element = (0018,0015); Name = BodyPartExamined; Sort = 1; Part = 1; },, {Element = (0028,0008); Name = NumberOfFrames; Sort = 1; Part = 1; },, {Element = (0028,0010); Name = Rows; Sort = 1; Part = 1; },, {Element = (0028,0011); Name = Columns; Sort = 1; Part = 1; },, {Element = (0020,0037); Name = ImageOrientationPatient; Sort = 1; Part = 1; Tolerance = 0.000005; },, {Element = (0018,1004); Name = PlateID; Sort = 1; Part = 1; },, {Element = (0018,1000); Name = DeviceSerialNumber; Sort = 1; Part = 1; },, {Element = (0028,0004); Name = PhotometricInterpretation; Sort = 1; Part = 1; },, {Element = (0028,0100); Name = BitsAllocated; Sort = 1; Part = 1; },, {Element = (0028,0102); Name = HighBit; Sort = 1; Part = 1; },, {Element = (0028,0103); Name = PixelRepresentation; Sort = 1; Part = 1; },, {Element = (0018,0020); Name = ScanningSequence; Sort = 1; Part = 1; },, {Element = (0018,0023); Name = MRAcquisitionType; Sort = 1; Part = 1; },, {Element = (0028,0030); Name = PixelSpacing; Sort = 1; Part = 1; Tolerance = 0.00001; },, {Element = (0054,0010); Name = EnergyWindowVector; Sort = 1; Part = 1; },, {Element = (0054,0030); Name = PhaseVector; Sort = 1; Part = 1; },, {Element = (0054,0060); Name = RRIntervalVector; Sort = 1; Part = 1; },, {Element = (0054,0050); Name = RotationVector; Sort = 1; Part = 1; },, {Element = (0028,1101); Name = RedPaletteColorLookupTableDescriptor; Sort = 1; Part = 1; },, {Element = (0028,1102); Name = GreenPaletteColorLookupTableDescriptor; Sort = 1; Part = 1; },, {Element = (0028,1103); Name = BluePaletteColorLookupTableDescriptor; Sort = 1; Part = 1; },, {Element = (0028,1201); Name = RedPaletteColorLookupTableData; Sort = 1; Part = 1; },, {Element = (0028,1202); Name = GreenPaletteColorLookupTableData; Sort = 1; Part = 1; },, {Element = (0028,1203); Name = BluePaletteColorLookupTableData; Sort = 1; Part = 1; },, {Element = (0020,000e); Name = SeriesInstanceUID; Sort = 1; SortCondition = "Modality = NM & !(FrameIncrementPointer = '(0054,0010)')" Part = 1; PartCondition = "Modality = NM & !(FrameIncrementPointer = '(0054,0010)')"; },, {Element = (0008,0018); Name = SOPInstanceUID; Sort = 1; SortCondition = "Modality = NM & NumberOfFrames > 1"; Part = 1; PartCondition = "Modality = NM & NumberOfFrames > 1"; },, {Element = (0008,0018); Name = SOPInstanceUID; Sort = 1; SortCondition = "Modality = CR | Modality = DR | Modality = MG | Modality = MX | Modality = RG"; Part = 1; PartCondition = "Modality = CR | Modality = DR | Modality = MG | Modality = MX | Modality = RG"; },, {Element = (0008,1090); Name = ManufacturerModelName; Sort = 1; Part = 1; },, {Element = (0008,1010); Name = StationName; Sort = 1; Part = 1; },, {Element = (0008,0070); Name = Manufacturer; Sort = 1; Part = 1; },, {Element = (0018,0020); Name = ScanningSequence; Sort = 1; Part = 1; },, {Element = (0018,0085); Name = ImagedNucleus; Sort = 1; Part = 1; },, {Element = (0018,0087); Name = MagneticFieldStrength; Sort = 1; Part = 1; },, {Element = (0018,1020); Name = SoftwareVersion; Sort = 1; Part = 1; },, {Element = (0018,5100); Name = PatientPosition; Sort = 1; Part = 1; },, {Element = (0062,000b); Name = ReferencedSegmentNumber; Sort = 1; SortCondition = "SOPClassUID = 1.2.840.10008.5.1.4.1.1.66.4"; Part = 1; PartCondition = "SOPClassUID = 1.2.840.10008.5.1.4.1.1.66.4"; },, {Element = (0020,000e); Name = SeriesInstanceUID; Sort = 1; SortCondition = "SOPClassUID = 1.2.840.10008.5.1.4.1.1.66.4"; Part = 1; PartCondition = "SOPClassUID = 1.2.840.10008.5.1.4.1.1.66.4"; },, {Element = (0020,0013); Name = InstanceNumber; Sort = 1; Part = 0; },, {Element = (0008,0030); Name = StudyTime; Sort = 1; Part = 0; },, {Element = (0020,0011); Name = SeriesNumber; Sort = 1; Part = 0; },, {Element = (0008,0021); Name = SeriesDate; Sort = 1; Part = 0; },, {Element = (0008,0031); Name = SeriesTime; Sort = 1; Part = 0; },, {Element = (0018,0081); Name = EchoTime; Sort = 1; Part = 0; },, {Element = (0018,0024); Name = SequenceName; Sort = 1; Part = 0; },, {Element = (0020,0012); Name = AcquisitionNumber; Sort = 1; Part = 0; },, {Element = (0018,0022); Name = ScanOptions; Sort = 1; Part = 0; },, {Element = (0008,0022); Name = AcquisitionDate; Sort = 1; Part = 0; },, {Element = (0008,0032); Name = AcquisitionTime; Sort = 1; Part = 0; },, {Element = (0008,0023); Name = ContentDate; Sort = 1; Part = 0; },, {Element = (0008,0033); Name = ContentTime; Sort = 1; Part = 0; },, {Element = (0020,0032); Name = ImagePositionPatient; Sort = 1; Part = 0; },, {Element = (0054,0020); Name = DetectorVector; Sort = 1; Part = 0; },, {Element = (0054,0100); Name = TimeSliceVector; Sort = 1; Part = 0; },, {Element = (0054,0070); Name = TimeSlotVector; Sort = 1; Part = 0; },, {Element = (0054,0080); Name = SliceVector; Sort = 1; Part = 0; },, {Element = (0054,0090); Name = AngularViewVector; Sort = 1; Part = 0; },, {Element = (0020,9157); Name = DimensionIndexValues; Sort = 1; Part = 0; }, )¶
Sets the sort/part configuration without a file.
Used if
Use file for sort/partis False.
Reset to Default¶
- name: resetSortPartToDefault, type: Trigger¶
When pressed, the
Direct Sort Part Configurationis reset to its default value.
Check Sort Part¶
- name: checkSortPart, type: Trigger¶
When pressed, the sort/part configuration specified in
Direct Sort Part ConfigurationorSort Part Config Fileis validated.
Had warnings¶
- name: checkedSortPartHadWarnings, type: Bool, persistent: no¶
Shows whether the sort/part configuration specified in
Direct Sort Part ConfigurationorSort Part Config Fileproduced any warnings when validated viaCheck Sort Partor during import.
Had errors¶
- name: checkedSortPartHadErrors, type: Bool, persistent: no¶
Shows whether the sort/part configuration specified in
Direct Sort Part ConfigurationorSort Part Config Fileproduced any errors when validated viaCheck Sort Partor during import.An empty sort/part file referenced by
Sort Part Config Fileis treated as an error.
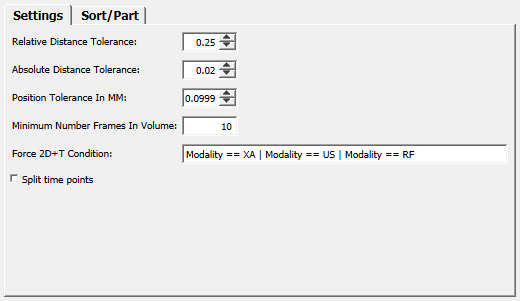
Relative Distance Tolerance¶
- name: relativeDistanceTolerance, type: Double, default: 0.25, minimum: 0¶
Sets a factor that is applied to the distances between previous frames in a block (after sorting) to obtain a tolerance value.
Additionally, the value of
Absolute Distance Toleranceis added. If the position of a frame differs by more than the resulting value from the expected position, a new volume is started.
Absolute Distance Tolerance¶
- name: absoluteDistanceTolerance, type: Double, default: 0.02, minimum: 0¶
Sets a value that is added to the tolerance described in
Relative Distance Tolerance.
Position Tolerance In MM¶
- name: positionToleranceInMM, type: Double, default: 0.0999, minimum: 0¶
Sets the minimum amount by which the position of a single frame should differ from the previous frame.
If the difference is too small, the frame is assumed to be at the same position, but at a different time point.
Minimum Number Frames In Volume¶
- name: minimumNumberFramesInVolume, type: Integer, default: 10, minimum: 1¶
Sets the minimum number of frames in a volume.
If a volume would be composed of fewer frames, the frames are imported as single frames instead.
Force 2D+T Condition¶
- name: force2DPlusTCondition, type: String, default: Modality == XA | Modality == US | Modality == RF¶
Sets a condition that describes when to enforce building a time series from frames instead of a 3D block.
The frames will have the same world position in MeVisLab in this case.
This is only applied if a time series is not generated otherwise, which might result in a 4D block instead.
Split Time Points¶
- name: splitTimePoints, type: Bool, default: FALSE¶
If checked, the creation of 4D images is prevented by splitting them into 3D volumes. Does not affect 2D+T images.
>¶
- name: selectNextItem, type: Trigger¶
When pressed, the next volume is selected in the patient tree.
<¶
- name: selectPreviousItem, type: Trigger¶
When pressed, the previous volume is selected in the patient tree.