FeatureSpaceMatrix¶
- MLModule¶
genre
authors
package
dll
definition
keywords
Purpose¶
The module FeatureSpaceMatrix calculates the feature space matrix from a fiber set which may be used for fiber clustering.
Windows¶
Default Panel¶
Input Fields¶
inputFiberSet¶
- name: inputFiberSet, type: MLBase¶
A FiberSet or FiberSetContainer.
Output Fields¶
outFeatureMatrix¶
- name: outFeatureMatrix, type: MLBase¶
The feature space matrix with dimension numObjects x numFeatures.
outFeatureWeightVector¶
- name: outFeatureWeightVector, type: MLBase¶
Feature weights used in
ClusteringAlgorithmsstored as a vector with dimension (numFeatures x numSegments).
Parameter Fields¶
Field Index¶
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Visible Fields¶
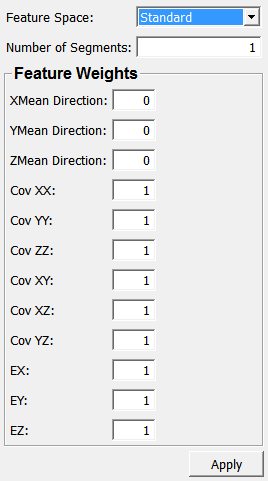
Feature Space¶
- name: featureSpace, type: Enum, default: Standard¶
Defines the feature space in which to work in.
Values:
Title |
Name |
Description |
|---|---|---|
Standard |
Standard |
Feature space corresponding to input fibers and selected feature weights. |
Feature Space 1 |
Feature Space 1 |
Not implemented: arbitrary feature spaces could be added in the future. |
Number of Segments¶
- name: numSegments, type: Integer, default: 1¶
Sets the number of target segments.
Each fiber is partitioned into the desired number of segments, independently of its length. Then each segment is treated as a short fiber and the whole feature space is calculated and the feature spaces combined (numFeatures x numSegments resulting columns).
However this procedure does not yield great results: the algorithms (in
ClusteringAlgorithms), which compute the affinity matrix , sometimes end up comparing two fibers starting from opposite ends. In addition a high dimensional feature space usually leads to many weights being of the same order of magnitude and therefore to a non-sparse affinity matrix and ambiguous clustering.
XMean Direction¶
- name: xMeanDirection, type: Float, default: 0¶
Sets the weighting of mean x-components.
YMean Direction¶
- name: yMeanDirection, type: Float, default: 0¶
Sets the weighting of mean y-components.
ZMean Direction¶
- name: zMeanDirection, type: Float, default: 0¶
Sets the weighting of mean z-components.
Cov XX¶
- name: covXX, type: Float, default: 1¶
Sets the weighting of cov(xx).
Cov YY¶
- name: covYY, type: Float, default: 1¶
Sets the weighting of cov(yy).
Cov ZZ¶
- name: covZZ, type: Float, default: 1¶
Sets the weighting of cov(zz).
Cov XY¶
- name: covXY, type: Float, default: 1¶
Sets the weighting of cov(xy).
Cov XZ¶
- name: covXZ, type: Float, default: 1¶
Sets the weighting of cov(xz).
Cov YZ¶
- name: covYZ, type: Float, default: 1¶
Sets the weighting of cov(yz).
EX¶
- name: eX, type: Float, default: 1¶
Sets the weighting of ex.
EY¶
- name: eY, type: Float, default: 1¶
Sets the weighting of ey.
EZ¶
- name: eZ, type: Float, default: 1¶
Sets the weighting of ez.
&Apply¶
- name: apply, type: Trigger¶
If pressed, the module computes anew.