| 6.6. C-Example using the C-API | ||
|---|---|---|
 | Chapter 6. The C-API |  |
The following section contains a small C example that creates an
ML module network for loading, filtering and saving an image. Note that
the libraries for MLUtilities,
MLLinearAlgebra, ML,
MLImageFile, MLGeometry1,
MLDicomTree_OFFIS and MLImageIO must
be available in binary search paths to run the program correctly. They
can normally be found in the installation directory of MeVisLab which is
usually available when working with the ML.
The example program implements the following operations:
Checking the number of command line parameters and the validity of the ML version,
Loading the libraries MLImageFile,
MLGeometry1, and
MLDicomTree_OFFIS to have all required modules
linked to the executable,
Creating an ImgLoad,
Resample3D, and
ImageSave module,
Setting input and output file names in
ImageLoad and
ImageSave module,
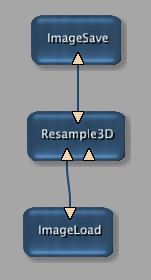
Connecting ImageLoad,
Resample3D, and
ImageSave module to a module pipeline,
and
Setting zoom parameter in Resample3D,
saving the result in a file by triggering
ImageSave, and checking the status field of
ImageSave.
Example 6.1. Using the C-API
// Simple ML program that initializes the library, loads the given // dataset, applies a resampling and writes the result back to disk. // // The input file can be any format supported by the MFL (MeVis File Library) now called MLImageIO, // including DICOM (.dcm), TIFF (.tif,.tiff) // // The output file is written as a DICOM/TIFF combination typically used by // MeVisLab (DICOM header + tiff data). #include "mlAPI.h" #include <stdio.h> #include <iostream>
int main(int argc, char* argv[])
{
// run only if enough arguments
if (argc > 5) {
// Extra char buffer
char buffer[4096]="\n";
std::cout << "imagefilter: loading " << argv[1] << std::endl;
std::cout << "imagefilter: output " << argv[2] << std::endl;
// Initialize the ML.
MLInit(ML_MAJOR_VERSION, ML_MAJOR_CAPI_VERSION, ML_CAPI_REVISION);
// Load additional image file and filter module libraries.
MLLoadLibrary("MLImageFile");
MLLoadLibrary("MLGeometry1");
// Also load a DICOM tree implementation to be able to load DICOM images.
MLLoadLibrary("MLDicomTree_OFFIS");
//--------------------------------------------
// Create modules
// Create an ImgLoad module.
mlModule* loader = MLCreateModuleFromName("ImgLoad");
// Create resample module.
mlModule* resample = MLCreateModuleFromName("Resample3D");
// Create an ImgSave module.
mlModule* writer = MLCreateModuleFromName("ImgSave");
//--------------------------------------------
// Setup file names
// Get the file name field of the loader.
mlField* loaderFilenameField = MLModuleGetField(loader,"filename");
// Set the file name field to the given command line argument
MLFieldSetValue(loaderFilenameField,argv[1]);
// Get the file name field of the writer.
mlField* writerFilenameField = MLModuleGetField(writer,"filename");
// Set the file name field to the given command line argument
MLFieldSetValue(writerFilenameField,argv[2]);
//--------------------------------------------
// Connect modules
// Get the output image field of the loader.
mlField* loaderOutput0 = MLModuleGetField(loader,"output0");
// Get the input image field of the resample module.
mlField* resampleInput0 = MLModuleGetField(resample,"input0");
// Connect input of resample to output of loader.
// Always connect input to output (destination to source) and not vice versa.
MLFieldConnectFrom(resampleInput0,loaderOutput0);
// Get the output image field of the resample module.
mlField* resampleOutput0 = MLModuleGetField(resample,"output0");
// Get the input image field of the writer module.
mlField* writerInput0 = MLModuleGetField(writer,"input0");
// Connect input of resample to output of loader.
MLFieldConnectFrom(writerInput0,resampleOutput0);
//--------------------------------------------
// Set zoom factor
// Get zoom factor field.
mlField* zoomField = MLModuleGetField(resample,"zoom");
// Concatenate arguments to form a vector string.
sprintf(buffer,"%s %s %s",argv[3],argv[4],argv[5]);
// Set vector string value to zoom field.
MLFieldSetValue(zoomField,buffer);
//--------------------------------------------
// Write image back to disk
// Get save field from writer.
mlField* saveField = MLModuleGetField(writer,"save");
// Touch the save trigger, this actually saves the image to disk.
std::cerr << "Starting image save..." << std::endl;
MLFieldTouch(saveField);
std::cerr << "...finished." << std::endl;
//--------------------------------------------
// Check if writing was ok
mlField* statusField = MLModuleGetField(writer,"status");
// Get value of status field into given buffer (maximum buffer size is also passed).
MLFieldGetValue(statusField, buffer, 4096);
std::cout << "Write status: " << buffer << std::endl;
} else {
std::cout << "Usage: imagefilter inputfile outputfile xscale yscale zscale" << std::endl;
}
return 0;
}This example called with the command line arguments
/demodata/Carotid1_MRA.small.dcm Carotid1_MRA.small.scaled.dcm 1 2 3
is comparable to the following module network and panels in MeVisLab:
 |
© 2025 MeVis Medical Solutions AG