WeightMatrixByGrid¶
- MLModule¶
genre
authors
package
dll
definition
keywords
Purpose¶
The module WeightMatrixByGrid calculates an affinity matrix from a fiber set using the fiber-grid method proposed in the paper Grid-Based Spectral Fiber Clustering.
Details¶
There are two natural ways to define an affinity matrix:
weight matrix: A high value of the element ij corresponds to a high affinity between i and j.
distance matrix: A high value of the element ij corresponds to a low affinity between i and j.
Each object of the class Matrix has a tag indicating the type of the matrix (e.g. distanceMatrix or weightMatrix).
The module ClusteringAlgorithms checks those tags and converts the type of a matrix if necessary.
Windows¶
Default Panel¶
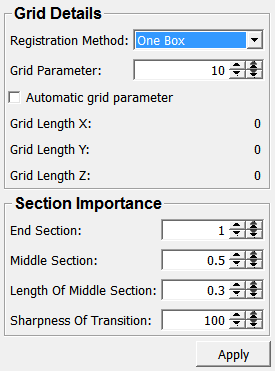
Input Fields¶
inputFiberSet¶
- name: inputFiberSet, type: MLBase¶
A FiberSet.
Output Fields¶
outMatrix¶
- name: outMatrix, type: MLBase¶
Affinity matrix as a normalized weight matrix.
outCurve¶
- name: outCurve, type: MLBase¶